Overview
Note
This page describes how a standard phyddle pipeline analysis is configured and how settings determine software behavior. Main sections: Configuration, Pipeline, Workspace, Formats, and Safe Usage. Visit Glossary to learn more about how phyddle defines different terms.
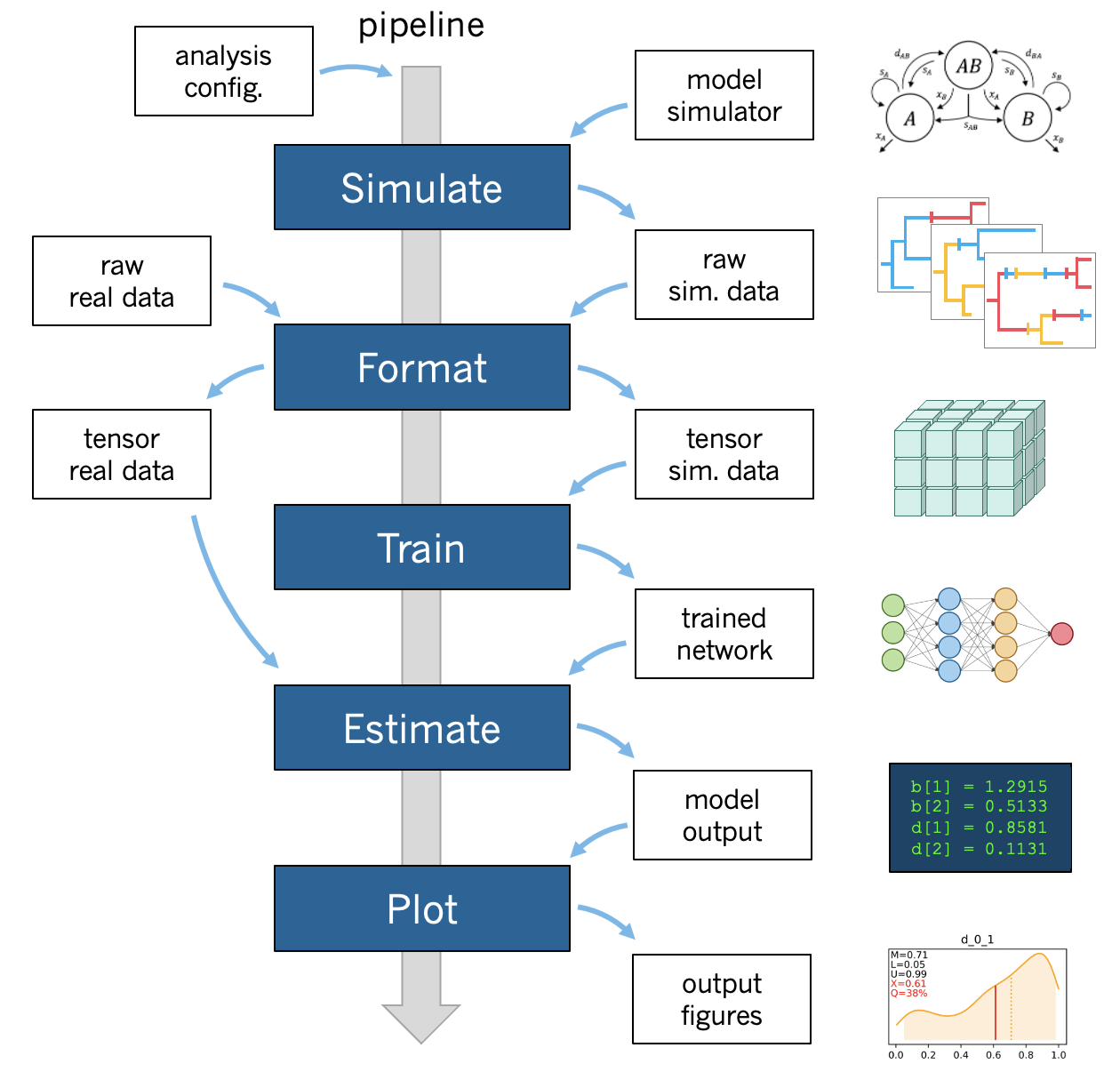
A phyddle pipeline analysis has five steps: Simulate, Format, Train, Estimate, and Plot. Standard analyses run all steps, in order, based on the analysis Configuration. Steps can also be run individually and/or multiple times under different settings and orders, which is useful for exploratory and advanced analyses.
All pipeline steps create output files, which are stored in the Workspace directories. All pipeline (except Simulate) also require input files corresponding to at least one other pipeline step. A full phyddle analysis for a project will automatically generate the input files for downstream pipeline steps and store them in a predictable project directory.
Users may also elect to use phyddle for only some steps in their analysis, and produce files for other steps by different means. For example, Format expects to format and combine large numbers of simulated datasets into tensor formats that can be used for supervised learning with neural networks. These simulated files can either be generated through phyddle with the Simulate step or outside of phyddle entirely.
Below is the standard Workspace directory structure that a standard
phyddle project would use. In this section, we assume the project dir is
./workspace/example:
Simulate
- input: None
- output: ./workspace/example/simulate # simulated datasets
Format
- input: ./workspace/example/simulate # simulated datasets
./workspace/example/empirical # empirical datasets
- output: ./workspace/example/format # formatted datasets
Train
- input: ./workspace/example/format # simulated training dataset
- output: ./workspace/example/train # trained network + train results
Estimate
- input: ./workspace/example/format # simulated test + empirical datasets
./workspace/example/train # trained network
- output: ./workspace/example/estimate # test + empirical results
Plot
- input: ./workspace/example/format # simulated training dataset
./workspace/example/train # trained network and output
./workspace/example/estimate # simulated + empirical estimates
- output: ./workspace/example/plot # analysis figures
Configuration
There are two ways to configure the settings of a phyddle analysis: through a config file or the command line. Command line settings take precedence over config file settings.
Note
The Appendix contains a Table of Settings that summarizes all available settings.
By file
The phyddle config file is a Python dictionary of analysis arguments (args)
that configure how phyddle pipeline steps behave. Because it’s a Python script,
you can write code within the config file to specify your analysis, if you find
that helpful. The below example defines settings into different blocks based on
which pipeline step first needs a given setting. However, any setting might be
used by different pipeline steps, so we concatenate all settings into a single
dictionary called args, which is then used by all pipeline steps. Settings
configured by file can be adjusted through the command line,
if desired.
Note
By default, phyddle assumes you want to use the config file called
config.py. Use a different config file by calling, e.g.
phyddle --cfg my_other_config.py
Note
phyddle maintains a number of example config files for different models
and simulation methods. These are organized as project subdirectories
within the ./workspace directory. For example,
./workspace/bisse_r/config.py simulates under a BiSSE model
with the R simulation script ./workspace/bisse_r/sim_bisse.R.
#==============================================================================#
# Config: Example phyddle config file #
# Authors: Michael Landis and Ammon Thompson #
# Date: 230804 #
# Description: Simple BiSSE model #
#==============================================================================#
args = {
#-------------------------------#
# Basic #
#-------------------------------#
'step' : 'SFTEP', # Pipeline step(s) defined with
# (S)imulate, (F)ormat, (T)rain,
# (E)stimate, (P)lot, or (A)ll
'verbose' : 'T', # Verbose output to screen?
#-------------------------------#
# Analysis #
#-------------------------------#
'use_parallel' : 'T', # Use parallelization? (recommended)
'num_proc' : -2, # Number of cores for multiprocessing
# (-N for all but N)
'use_cuda' : 'T', # Use CUDA parallelization?
# (recommended; requires Nvidia GPU)
#-------------------------------#
# Workspace #
#-------------------------------#
'dir' : './', # Base directory for all step directories
'prefix' : 'out', # Prefix for all output unless step prefix given
#-------------------------------#
# Simulate #
#-------------------------------#
'sim_command' : f'Rscript ./sim_bisse.R', # Simulation command to run single
# job (see documentation)
'sim_logging' : 'verbose', # Simulation logging style
'start_idx' : 0, # Start index for simulated training replicates
'end_idx' : 1000, # End index for simulated training replicates
'sim_batch_size' : 10, # Number of replicates per simulation command
#-------------------------------#
# Format #
#-------------------------------#
'encode_all_sim' : 'T', # Encode all simulated replicates into tensor?
'num_char' : 1, # Number of characters
'num_states' : 2, # Number of states per character
'min_num_taxa' : 10, # Minimum number of taxa allowed when formatting
'max_num_taxa' : 500, # Maximum number of taxa allowed when formatting
'downsample_taxa' : 'uniform', # Downsampling strategy taxon count
'tree_width' : 500, # Width of phylo-state tensor
'tree_encode' : 'extant', # Encoding strategy for tree
'brlen_encode' : 'height_brlen', # Encoding strategy for branch lengths
'char_encode' : 'integer', # Encoding strategy for character data
'param_est' : { # Unknown model parameters to estimate
'log10_birth_1' : 'num',
'log10_birth_2' : 'num',
'log10_death' : 'num',
'log10_state_rate' : 'num',
'model_type' : 'cat',
'root_state' : 'cat'
],
'param_data' : { # Known model parameters to treat as aux. data
'sample_frac' : 'num'
},
'char_format' : 'csv', # File format for character data
'tensor_format' : 'hdf5', # File format for training example tensors
#-------------------------------#
# Train #
#-------------------------------#
'num_epochs' : 20, # Number of training epochs
'trn_batch_size' : 2048, # Training batch sizes
'prop_test' : 0.05, # Proportion of data used as test examples
# (to assess trained network performance)
'prop_val' : 0.05, # Proportion of data used as validation examples
# (to diagnose network overtraining)
'prop_cal' : 0.2, # Proportion of data used as calibration examples
# (to calibrate CPIs)
'cpi_coverage' : 0.95, # Expected coverage percent for calibrated
# prediction intervals (CPIs)
'cpi_asymmetric' : 'T', # Use asymmetric (True) or symmetric (False)
# adjustments for CPIs?
'loss' : 'mae', # Loss function for optimization
'optimizer' : 'adam', # Method used for optimizing neural network
'phy_channel_plain' : [64, 96, 128], # Output channel sizes for plain convolutional
# layers for phylogenetic state input
'phy_channel_stride' : [64, 96], # Output channel sizes for stride convolutional
# layers for phylogenetic state input
'phy_channel_dilate' : [32, 64], # Output channel sizes for dilate convolutional
# layers for phylogenetic state input
'aux_channel' : [128, 64, 32], # Output channel sizes for dense layers for
# auxiliary data input
'lbl_channel' : [128, 64, 32], # Output channel sizes for dense layers for
# label outputs
'phy_kernel_plain' : [3, 5, 7], # Kernel sizes for plain convolutional layers
# for phylogenetic state input
'phy_kernel_stride' : [7, 9], # Kernel sizes for stride convolutional layers
# for phylogenetic state input
'phy_kernel_dilate' : [3, 5], # Kernel sizes for dilate convolutional layers
# for phylogenetic state input
'phy_stride_stride' : [3, 6], # Stride sizes for stride convolutional layers
# for phylogenetic state input
'phy_dilate_dilate' : [3, 5], # Dilation sizes for dilate convolutional layers
# for phylogenetic state input
#-------------------------------#
# Estimate #
#-------------------------------#
# not currently used
#-------------------------------#
# Plot #
#-------------------------------#
'plot_train_color' : 'blue', # Plotting color for training data elements
'plot_label_color' : 'orange', # Plotting color for training label elements
'plot_test_color' : 'purple', # Plotting color for test data elements
'plot_val_color' : 'red', # Plotting color for validation data elements
'plot_aux_color' : 'green', # Plotting color for auxiliary data elements
'plot_emp_color' : 'black', # Plotting color for empirical elements
'plot_num_scatter' : 50, # Number of examples in scatter plot
'plot_min_emp' : 5, # Minimum number of empirical datasets to plot densities
'plot_num_emp' : 10 # Number of empirical results to plot
}
Via command line
Settings applied through a config file can be overwritten
by setting options when running phyddle from the command line. The names of
settings are the same for the command line options and in the config file.
Using command line options makes it easy to adjust the behavior of pipeline
steps without needing to edit the config file. List all settings that can be
adjusted with the command line using the --help option:
usage: phyddle [-h] [-c] [-s] [-v] [--make_cfg ] [--save_proj ] [--load_proj ]
[--clean_proj ] [--save_num_sim] [--save_train_fmt]
[--output_precision] [--use_parallel] [--use_cuda] [--num_proc]
[--no_emp] [--no_sim] [--dir] [--sim_dir] [--emp_dir]
[--fmt_dir] [--trn_dir] [--est_dir] [--plt_dir] [--log_dir]
[--prefix] [--sim_prefix] [--emp_prefix] [--fmt_prefix]
[--trn_prefix] [--est_prefix] [--plt_prefix] [--sim_command]
[--sim_logging {clean,compress,verbose}] [--start_idx]
[--end_idx] [--sim_more] [--sim_batch_size] [--encode_all_sim]
[--num_char] [--num_states] [--min_num_taxa] [--max_num_taxa]
[--downsample_taxa {uniform}] [--tree_width]
[--tree_encode {extant,serial}]
[--brlen_encode {height_only,height_brlen}]
[--char_encode {one_hot,integer,numeric}] [--param_est]
[--param_data] [--char_format {csv,nexus}]
[--tensor_format {csv,hdf5}] [--save_phyenc_csv] [--num_epochs]
[--num_early_stop] [--trn_batch_size] [--prop_test]
[--prop_val] [--prop_cal] [--cpi_coverage] [--cpi_asymmetric]
[--loss_numerical {mse,mae}] [--optimizer {adam}]
[--log_offset] [--phy_channel_plain] [--phy_channel_stride]
[--phy_channel_dilate] [--aux_channel] [--lbl_channel]
[--phy_kernel_plain] [--phy_kernel_stride]
[--phy_kernel_dilate] [--phy_stride_stride]
[--phy_dilate_dilate] [--plot_train_color] [--plot_test_color]
[--plot_val_color] [--plot_label_color] [--plot_aux_color]
[--plot_emp_color] [--plot_num_scatter] [--plot_min_emp]
[--plot_num_emp] [--plot_pca_noise]
Software to fiddle around with deep learning for phylogenetic models
options:
-h, --help show this help message and exit
-c, --cfg Config file name
-s, --step Pipeline step(s) defined with (S)imulate, (F)ormat,
(T)rain, (E)stimate, (P)lot, or (A)ll
-v, --verbose Verbose output to screen?
--make_cfg Write default config file
--save_proj Save and zip a project for sharing
--load_proj Unzip a shared project
--clean_proj Remove step directories for a project
--save_num_sim Number of simulated examples to save with --save_proj
--save_train_fmt Save formatted training examples with --save_proj?
(not recommended)
--output_precision Number of digits (precision) for numbers in output
files
--use_parallel Use parallelization? (recommended)
--use_cuda Use CUDA parallelization? (recommended; requires
Nvidia GPU)
--num_proc Number of cores for multiprocessing (-N for all but N)
--no_emp Disable Format/Estimate steps for empirical data?
--no_sim Disable Format/Estimate steps for simulated data?
--dir Parent directory for all step directories unless step
directory given
--sim_dir Directory for raw simulated data
--emp_dir Directory for raw empirical data
--fmt_dir Directory for tensor-formatted data
--trn_dir Directory for trained networks and training output
--est_dir Directory for new datasets and estimates
--plt_dir Directory for plotted results
--log_dir Directory for logs of analysis metadata
--prefix Prefix for all output unless step prefix given
--sim_prefix Prefix for raw simulated data
--emp_prefix Prefix for raw empirical data
--fmt_prefix Prefix for tensor-formatted data
--trn_prefix Prefix for trained networks and training output
--est_prefix Prefix for estimate results
--plt_prefix Prefix for plotted results
--sim_command Simulation command to run single job (see documentation)
--sim_logging {clean,compress,verbose}
Simulation logging style
--start_idx Start replicate index for simulated training dataset
--end_idx End replicate index for simulated training dataset
--sim_more Add more simulations with auto-generated indices
--sim_batch_size Number of replicates per simulation command
--encode_all_sim Encode all simulated replicates into tensor?
--num_char Number of characters
--num_states Number of states per character
--min_num_taxa Minimum number of taxa allowed when formatting
--max_num_taxa Maximum number of taxa allowed when formatting
--downsample_taxa {uniform}
Downsampling strategy taxon count
--tree_width Width of phylo-state tensor
--tree_encode {extant,serial}
Encoding strategy for tree
--brlen_encode {height_only,height_brlen}
Encoding strategy for branch lengths
--char_encode {one_hot,integer,numeric}
Encoding strategy for character data
--param_est Model parameters and variables to estimate
--param_data Model parameters and variables treated as data
--char_format {csv,nexus}
File format for character data
--tensor_format {csv,hdf5}
File format for training example tensors
--save_phyenc_csv Save encoded phylogenetic tensor encoding to csv?
--num_epochs Number of training epochs
--num_early_stop Number of consecutive validation loss gains before
early stopping
--trn_batch_size Training batch sizes
--prop_test Proportion of data used as test examples (assess
trained network performance)
--prop_val Proportion of data used as validation examples
(diagnose network overtraining)
--prop_cal Proportion of data used as calibration examples
(calibrate CPIs)
--cpi_coverage Expected coverage percent for calibrated prediction
intervals (CPIs)
--cpi_asymmetric Use asymmetric (True) or symmetric (False) adjustments
for CPIs?
--loss_numerical {mse,mae}
Loss function for real value estimates
--optimizer {adam} Method used for optimizing neural network
--log_offset Offset size c when taking ln(x+c) for zero-valued
variables
--phy_channel_plain Output channel sizes for plain convolutional layers
for phylogenetic state input
--phy_channel_stride
Output channel sizes for stride convolutional layers
for phylogenetic state input
--phy_channel_dilate
Output channel sizes for dilate convolutional layers
for phylogenetic state input
--aux_channel Output channel sizes for dense layers for auxiliary
data input
--lbl_channel Output channel sizes for dense layers for label
outputs
--phy_kernel_plain Kernel sizes for plain convolutional layers for
phylogenetic state input
--phy_kernel_stride Kernel sizes for stride convolutional layers for
phylogenetic state input
--phy_kernel_dilate Kernel sizes for dilate convolutional layers for
phylogenetic state input
--phy_stride_stride Stride sizes for stride convolutional layers for
phylogenetic state input
--phy_dilate_dilate Dilation sizes for dilate convolutional layers for
phylogenetic state input
--plot_train_color Plotting color for training data elements
--plot_test_color Plotting color for test data elements
--plot_val_color Plotting color for validation data elements
--plot_label_color Plotting color for label elements
--plot_aux_color Plotting color for auxiliary data elements
--plot_emp_color Plotting color for empirical elements
--plot_num_scatter Number of examples in scatter plot
--plot_min_emp Minimum number of empirical datasets to plot densities
--plot_num_emp Number of empirical results to plot
--plot_pca_noise Scale of Gaussian noise to add to PCA plot
Note: the step setting controls which steps should be applied.
Each pipeline step is represented by a capital letter:
S for Simulate, F for Format, T for Train,
E for Estimate, P for Plot, and A for all steps.
For example, the following two commands are equivalent
phyddle --step A
phyddle -s SFTEP
whereas calling
phyddle -s SF
commands phyddle to perform the Simulate and Format steps, but not the Train, Estimate, and Plot steps.
Pipeline
A standard phyddle analysis runs five steps – Simulate, Format, Train, Estimate, and Plot – in order.
Step directories
In general, each phyddle analysis will store all work within a single project directory. Work from each step, however, is stored into different subdirectories.
Customizing the input and output directories among steps allows users to quickly explore alternative pipeline designs while leaving previous pipeline results in place.
The project directory can be set using dir. During analysis, phyddle will
create subdirectories for each step using default names, as needed. For example,
if dir is set to the local directory ./, then a full phyddle analysis
would use the following directories for the analysis:
./simulate # default sim_dir
./empirical # default emp_dir
./format # default fmt_dir
./train # default trn_dir
./estimate # default est_dir
./plot # default plt_dir
./log # default log_dir
Individual step directories can be overriden with custom directory locations.
For example, setting dir to ./ but setting emp_dir to
/Users/mlandis/datasets/viburnum and plt_dir to
/Users/mlandis/projects/viburnum/results would cause
phyddle to use the following directories:
./simulate # default sim_dir
/Users/mlandis/datasets/viburnum # custom emp_dir
./format # default fmt_dir
./train # default trn_dir
./estimate # default est_dir
/Users/mlandis/projects/viburnum/results # custom plt_dir
./log # default log_dir
Step prefixes
Standard phyddle analyses assume that the files generated by each pipeline
step begin with the filename prefix 'out'.
The filename prefix for all pipeline steps can be changed using the prefix
settings. Changing the filename prefix allows you to generate alternative
pipeline filesets without overwriting previous results.
As with the pipeline directory settings (above), prefixes for individual pipeline steps can be overridden with custom prefixes. This allows you to compare pipeline performance using different settings, while saving previous work. For example,
phyddle -c config.py \ # load config
-s TE \ # run Train and Estimate steps
--prefix new \ # T & E output has prefix 'new'
--fmt_prefix out \ # Format input has prefix 'out'
--num_epochs 50 \ # Train for 50 epochs
--trn_batch_size 4096 # Use batch sizes of 4096 samples
By default the Format and Estimate steps run in a greedy manner,
against the simulated datasets identified by dir (or sim_dir) and
prefix (or sim_prefix), and against the empirical datasets identified
by dir (or emp_dir) and prefix (or emp_prefix), should those
datasets exist.
Setting --no_sim during a command-line run will instruct phyddle to skip
the Format and Estimate steps for the simulated datasets (i.e. the train and
test datasets).
Setting --no_emp during a command-line run will instruct phyddle to skip
the Format and Estimate steps for the empirical datasets.
In particular, the --no_sim flag in particular is useful when you only
need to format new empirical datasets, but do not need to reformat existing
simulated (i.e. training/test) datasets. The flag helps eliminate redundant
formatting tasks during pipeline development.
Simulate
Simulate instructs phyddle to simulate your training dataset. Any simulator that can be called from command-line can be used to generate training datasets with phyddle. This allows researchers to use their favorite simulator with phyddle for phylogenetic modeling tasks.
As a worked example, suppose we have an R script called sim_bisse.R containing
the following code
#!/usr/bin/env Rscript
library(castor)
library(ape)
# disable warnings
options(warn = -1)
# example command string to simulate for "sim.1" through "sim.10"
# cd ~/projects/phyddle/workspace/example
# Rscript sim_bisse.R ./simulate example 1 10
# arguments
args = commandArgs(trailingOnly = TRUE)
out_path = args[1]
out_prefix = args[2]
start_idx = as.numeric(args[3])
batch_size = as.numeric(args[4])
rep_idx = start_idx:(start_idx+batch_size-1)
num_rep = length(rep_idx)
get_mle = FALSE
# filesystem
tmp_fn = paste0(out_path, "/", out_prefix, ".", rep_idx) # sim path prefix
phy_fn = paste0(tmp_fn, ".tre") # newick file
dat_fn = paste0(tmp_fn, ".dat.csv") # csv of data
lbl_fn = paste0(tmp_fn, ".labels.csv") # csv of labels (e.g. params)
# dataset setup
num_states = 2
tree_width = 500
label_names = c(paste0("log10_birth_",1:num_states),
"log10_death", "log10_state_rate", "sample_frac")
# simulate each replicate
for (i in 1:num_rep) {
# set RNG seed
set.seed(rep_idx[i])
# rejection sample
num_taxa = 0
while (num_taxa < 10) {
# simulation conditions
max_taxa = runif(1, 10, 5000)
max_time = runif(1, 1, 100)
sample_frac = 1.0
if (max_taxa > tree_width) {
sample_frac = tree_width / max_taxa
}
# simulate parameters
Q = get_random_mk_transition_matrix(num_states, rate_model="ER",
max_rate=0.1)
birth = runif(num_states, 0, 1)
death = min(birth) * runif(1, 0, 1.0)
death = rep(death, num_states)
parameters = list(
birth_rates=birth,
death_rates=death,
transition_matrix_A=Q
)
# simulate tree/data
res_sim = simulate_dsse(
Nstates=num_states,
parameters=parameters,
sampling_fractions=sample_frac,
max_extant_tips=max_taxa,
max_time=max_time,
include_labels=T,
no_full_extinction=T)
# check if tree is valid
num_taxa = length(res_sim$tree$tip.label)
}
# save tree
tree_sim = res_sim$tree
write.tree(tree_sim, file=phy_fn[i])
# save data
state_sim = res_sim$tip_states - 1
df_state = data.frame(taxa=tree_sim$tip.label, data=state_sim)
write.csv(df_state, file=dat_fn[i], row.names=F, quote=F)
# save learned labels (e.g. estimated data-generating parameters)
label_sim = c(birth[1], birth[2], death[1], Q[1,2], sample_frac)
label_sim = log(label_sim, base=10)
names(label_sim) = label_names
df_label = data.frame(t(label_sim))
write.csv(df_label, file=lbl_fn[i], row.names=F, quote=F)
}
# done!
quit()
This particular script has a few important features. First, the simulator is
entirely responsible for simulating the dataset. Second, the script assumes it
will be provided runtime arguments (args`) to generate filenames and to
determine how many simulated datasets will be generated when the script is run
(more details in next paragraph). Third, output for the Newick string is stored
into a .tre file, for the character matrix data into a .dat.csv file,
and for the training labels into a comma-separated .csv file. Visit
Input datasets to learn more about input format requirements.
Now that we understand the script, we need to configure phyddle to call it
properly. This is done by setting the sim_command argument equal to a
command string of the form MY_COMMAND [MY_COMMAND_ARGUMENTS]. During
simulation, phyddle executes the command string against different filepath
locations. More specifically, phyddle will execute the command
MY_COMMAND [MY_COMMAND_ARGUMENTS] [SIM_DIR] [SIM_PREFIX], where SIM_DIR
is the path to the directory locating the individual simulated datasets, and
SIM_PREFIX is a common prefix shared by individual simulation files. As
part of the Simulate step, phyddle will execute the command string to generate
the complete simulated dataset of replicated training examples.
In this case, we assume that sim_bisse.R is an R script that is located in the same directory as config.py and can be executed using the Rscript command. The correct sim_command value to run this script is:
'sim_command' : 'Rscript ./sim_bisse.R'
Assuming sim_dir = './simulate', sim_prefix = 'sim'
sim_batch_size = 10, phyddle will execute the commands during simulation
Rscript sim_one.R ./simulate/ sim 0 10
Rscript sim_one.R ./simulate/ sim 10 10
Rscript sim_one.R ./simulate/ sim 20 10
...
for every replication index between start_idx and end_idx in
increments of sim_batch_size, where the R script itself is responsible
for generating the sim_batch_size replicates per batch. In fact,
executing Rscript sim_bisse.R ./simulate/ sim 1 10
from terminal is an ideal way to validate that your custom simulator is
compatible with the phyddle requirements.
Format
Format converts simulated and/or empirical data for a project into a tensor format that phyddle uses to train neural networks in the Train step. Format performs two main tasks:
Encode all individual raw datasets in the simulate and empirical project directory into individual tensor representations
Combines all the individual tensors into larger, singular tensors that can be processed by the neural network
For each example, Format encodes the raw data into two input tensors and one output tensor:
One input tensor is the phylogenetic-state tensor. Loosely speaking, these tensors contain information associated with clades across rows and information about relevant branch lengths and states per taxon across columns. The phylogenetic-state tensors used by phyddle are based on the compact bijective ladderized vector (CBLV) format of Voznica et al. (2022) and the compact diversity-reordered vector (CDV) format of Lambert et al. (2022) that incorporates tip states (CBLV+S and CDV+S) using the technique described in Thompson et al. (2022). Visit Tensor formats to learn more.
The second input is the auxiliary data tensor. This tensor contains summary statistics for the phylogeny and character data matrix and “known” parameters for the data generating process. Visit Auxiliary data to learn more.
The output tensor reports labels that are generally unknown data generating parameters to be estimated using the neural network. Depending on the estimation task, all or only some model parameters might be treated as labels for training and estimation.
For most purposes within phyddle, it is safe to think of a tensor as an n-dimensional array, such as a 1-d vector or a 2-d matrix. The tensor encoding ensures training examples share a standard shape (e.g. numbers of rows and columns) that helps the neural network to detect predictable data patterns. Learn more about the formats of phyddle tensors on the Tensor Formats page.
During tensor-encoding, Format processes the tree, data matrix, and
model parameters for each replicate. This is done in parallel, when the setting
use_parallel is set to True. Simulated data are processed using CBLV+S
format if tree_encode is set to 'serial'. If tree_encode is set to
'extant' then all non-extant taxa are pruned, saved as pruned.tre, then
encoded using CDV+S. Standard CBLV+S and CDV+S formats are used when
brlen_encode is 'height_only', while additional branch length
information is added as rows when brlen_encode is set to
'height_brlen'. Each tree is then encoded into a phylogenetic-state tensor
with a maximum of tree_width sampled taxa. Trees that contain more taxa are
downsampled to tree_width taxa. The number of taxa in each original dataset
is recorded in the summary statistics, allowing the trained network to
make estimates on trees that are larger or smaller than th exact tree_width
size.
The phylogenetic-state tensors and auxiliary data tensors are then created. If
save_phyenc_csv is set, then individual csv files are saved for each
dataset, which is especially useful for formatting new empirical datasets into
an accepted phyddle format.
The param_est setting identifies which “unknown”
parameters in the labels tensor you want to treat as downstream estimation
targets. The param_data setting identifies which of those parameters you
want to treat as “known” auxiliary data.
# Settings in config.py
# "unknown" parameters to estimate
'param_est' : {
'log10_birth_rate' : 'num',
'log10_death_rate' : 'num',
'log10_transition_matrix' : 'num',
'model_type' : 'cat',
'root_state' : 'cat'
}
# "known" parameters to use as auxiliary data
'param_data' : {
'sample_frac' : 'num'
}
Information for param_est and param_data are each stored as dictionaries.
The keys are the names of the parameters (labels) generated by the Simulate
step. The values are the data types of the parameters. Data types may be either
'num' for numerical parameters, or 'cat' for categorical parameters.
Note
Numerical parameters have ordered values that are negative, positive, or zero. We recommend that you transform bounded numerical parameters into unbounded values for use with phyddle. For example, although an evolutionary rate parameter is non-negative, the log of that rate can be negative, positive, or zero.
Note
Categorical parameters have unordered values. They are encoded using
base-0 sequential integers. For example, the nucleotides for an ancestral
state estimation task would use 0, 1, 2, 3 to represent A, C, G, T.
Lastly, Format creates a test dataset containing proportion test_prop of
all simulated examples, and a second training dataset that contains all
remaining 1.0 - test_prop examples.
Formatted tensors are then saved to disk either in simple comma-separated
value format or in a compressed HDF5 format. For example, suppose we set
fmt_dir to './format', fmt_prefix to 'out',
and tree_encode to 'serial'. If we set tensor_format == 'hdf5',
it produces:
./format/out.empirical.hdf5
./format/out.test.hdf5
./format/out.train.hdf5
or if tensor_format == 'csv':
./format/out.empirical.aux_data.csv
./format/out.empirical.labels.csv
./format/out.empirical.phy_data.csv
./format/out.test.aux_data.csv
./format/out.test.labels.csv
./format/out.test.phy_data.csv
./format/out.train.aux_data.csv
./format/out.train.labels.csv
./format/out.train.phy_data.csv
Format behaves the same way for simulated vs. empirical datasets,
except in two key ways. First, simulated datasets will be split into datasets
used to train the neural network and test its accuracy (in proportions defined
by test_prop), whereas empirical datasets are left whole. Second, simulated
datasets will contain labels for all data-generating parameters, meaning both
the “unknown” parameters that we want to estimate and the “known” parameters
that contribute to the data-generating process, but could be measured in the
real world. For example, the birth rate might be an “unknown” parameter we want
to estimate, while the missing proportion of species is a “known” parameter
that we can provide the network if we know e.g. only 10% of described
plant species are in the dataset.
When searching for empirical and simulated datasets, Format uses
emp_dir and sim_dir to locate the datasets. The emp_prefix and
sim_prefix settings are used to identify the datasets. Format
assumes that empirical datasets follow the naming pattern of
<prefix>.<rep_idx>.<ext> described for Simulate. For example,
setting emp_dir to '../dnds/empirical' and emp_prefix
to 'mammal_gene' will cause Format to search for files with
these names:
../dnds/empirical/mammal_gene.1.tre
../dnds/empirical/mammal_gene.1.dat.csv
../dnds/empirical/mammal_gene.1.labels.csv # if using known params
../dnds/empirical/mammal_gene.2.tre
../dnds/empirical/mammal_gene.2.dat.csv
../dnds/empirical/mammal_gene.2.labels.csv # if using known params
...
Using the --no_emp or --no_sim flags will instruct Format to
skip processing the empirical and simulated datasets, respectively. In
addition, Format will report that it is skipping the empirical and
simulated datasets if they do not exist.
Once complete, the formatted files can then be processed by the Train step and Estimate steps.
Train
Train builds a neural network and trains it to make model-based estimates using the simulated training example tensors compiled by the Format step.
The Train step performs six main tasks:
Load the input training example tensor.
Shuffle the input tensor and split it into training, test, validation, and calibration subsets.
Build and configure the neural network
Use supervised learning to train neural network to make accurate estimates (predictions)
Record network training performance to file
Save the trained network to file
Each network is trained for one set of prediction tasks for the exact model
as specified for the Simulate step. Each network is trained to
expect a specific set of Format settings (see above).
Important Format settings include tree_width, num_char,
num_states, char_encode, tree_encode, brlen_encode,
param_est, and param_known.
When the training dataset is read in, its examples are randomly shuffled by
replicate index. It then sets aside some examples for a validation dataset
(prop_val) and others for a calibration dataset (prop_cal). Note,
the Format step will have previously set aside some proportion of test
number of examples (prop_test) to measure final network accuracy
during the later Estimate step. The prop_val and prop_cal
are themselves proportions of the 1.0 - prop_test training example
proportion.
phyddle uses PyTorch <https://pytorch.org/> to build and train the network. The phylogenetic-state tensor is processed by convolutional and pooling layers, while the auxiliary data is processed by dense layers. All input layers are concatenated then pushed into three branches terminating in output layers to produce point estimates and upper and lower estimation intervals. Here is a simplified schematic of the network architecture:
Simplified network architecture:
Phylo. Data Aux. Data
Input Input
| |
.-------------+-------------. |
v v v v
Conv1D-plain Conv1D-dilate Conv1D-stride Dense
+ Pool + Pool + Pool |
. | | |
`-------------+----+---------+-----------'
|
v
Concat
+ Dense
|
.-----------+------+-----+-----------.
v v v v
Lower Point Upper Categ.
quantile estimate quantile estimates
The number of layers (depth) and nodes (width) can be controlled through the
phy_channel_plain, phy_channel_stride, phy_channel_dilate,
aux_channel, and lbl_channel settings. For example, the default
value for lbl_channel is [128, 64, 32],, which creates three dense
layers with 128, 64, and 32 output channels, in that order. The default value
for phy_channel_plain is [64, 96, 128], which creates three
sets of convolutional and average-pooling layers with 64, 96, and 128
output channels. Stride, dilation, and sizes for convolutional kernels can
be adjusted using phy_kernel_plain, phy_kernel_stride,
phy_kernel_dilate, phy_stride_stride, and phy_dilate_dilate.
Parameter point estimates use a loss function (e.g. loss_numerical
set to 'mse'). Lower and upper quantile estimates for numerical labels
are hard-coded to use a pinball loss function. Categorical label estimates are
hard-coded to use the cross-entropy loss function (one per cat. label).
Most layers share the the activation function defined by activation_func,
which uses rectified linear units (ReLUs) by default. The output channels
for each categorical variable uses a softmax activation function.
Calibrated prediction intervals (CPIs) are estimated using the conformalized
quantile regression technique of Romano et al. (2019). CPIs target a
particular estimation interval, e.g. set cpi_coverage to 0.80 so
80% of test estimations are expected contain the true simulating value.
More accurate CPIs can be obtained using two-sided conformalized quantile
regression by setting cpi_asymmetric to True, though this often
requires larger numbers of calibration examples, determined through
prop_cal.
The network is trained iteratively for num_epoch training cycles using
batch stochastic gradient descent, with batch sizes given by trn_batch_size.
Different optimizers can be used to update network weight and bias
parameters (e.g. optimizer == 'adam'). The initial learning rate can be
adjusted with learning_rate. Network performance is also
evaluated against a validation dataset, which contains prop_val of
all training examples, and not used for minimizing the loss function.
To prevent overtraining, phyddle will terminate training if the validation
loss does not improve for num_early_stop consecutive epochs (see
Safe Usage).
Training is automatically parallelized using CPUs and GPUs, dependent on
how Tensorflow was installed and system hardware. Output files are stored
in the directory assigned to trn_dir.
Estimate
Estimate loads the simulated and empirical test datasets created by
Format stored in fmt_dir with prefix fmt_prefix. For example,
if fmt_dir == './format', fmt_prefix == 'out',
and tensor_format == 'hdf5' then Estimate will process the
following files, if they exist:
./out.test.hdf5
./out.empirical.hdf5
This step then loads a pretrained network for a given tree_width and
uses it to estimate parameter values and calibrated prediction intervals
(CPIs) for both the empirical dataset and the (simulated) test dataset.
Estimates are then stored as separate files into the est_dir directory.
Using the --no_emp or --no_sim flags will instruct Estimate to
skip processing the empirical and simulated datasets, respectively. In
addition, Estimate will report that it is skipping the empirical and
simulated datasets if they do not exist.
When Estimate is involves empirical data, the step will report any input datasets our output estimates that have unusually extreme values relative to the training dataset. This is useful for identifying out-of-distribution errors (see Safe Usage).
Plot
Plot collects all results from the Format, Train, and Estimate steps to compile a set of useful figures, listed below. When results from Estimate are available, the step will integrate it into other figures to contextualize where that input dataset and estimated labels fall with respect to the training dataset.
Importantly, Plot will report instances where training, test, and empirical input datasets and output estimates are unusually different. This informs users of poor network behavior (see Safe Usage).
Plots are stored within plot_dir.
Colors for plot elements can be modified with plot_train_color,
plot_label_color, plot_test_color, plot_val_color,
plot_aux_color, and plot_est_color using hex codes or common color
names supported by Matplotlib.
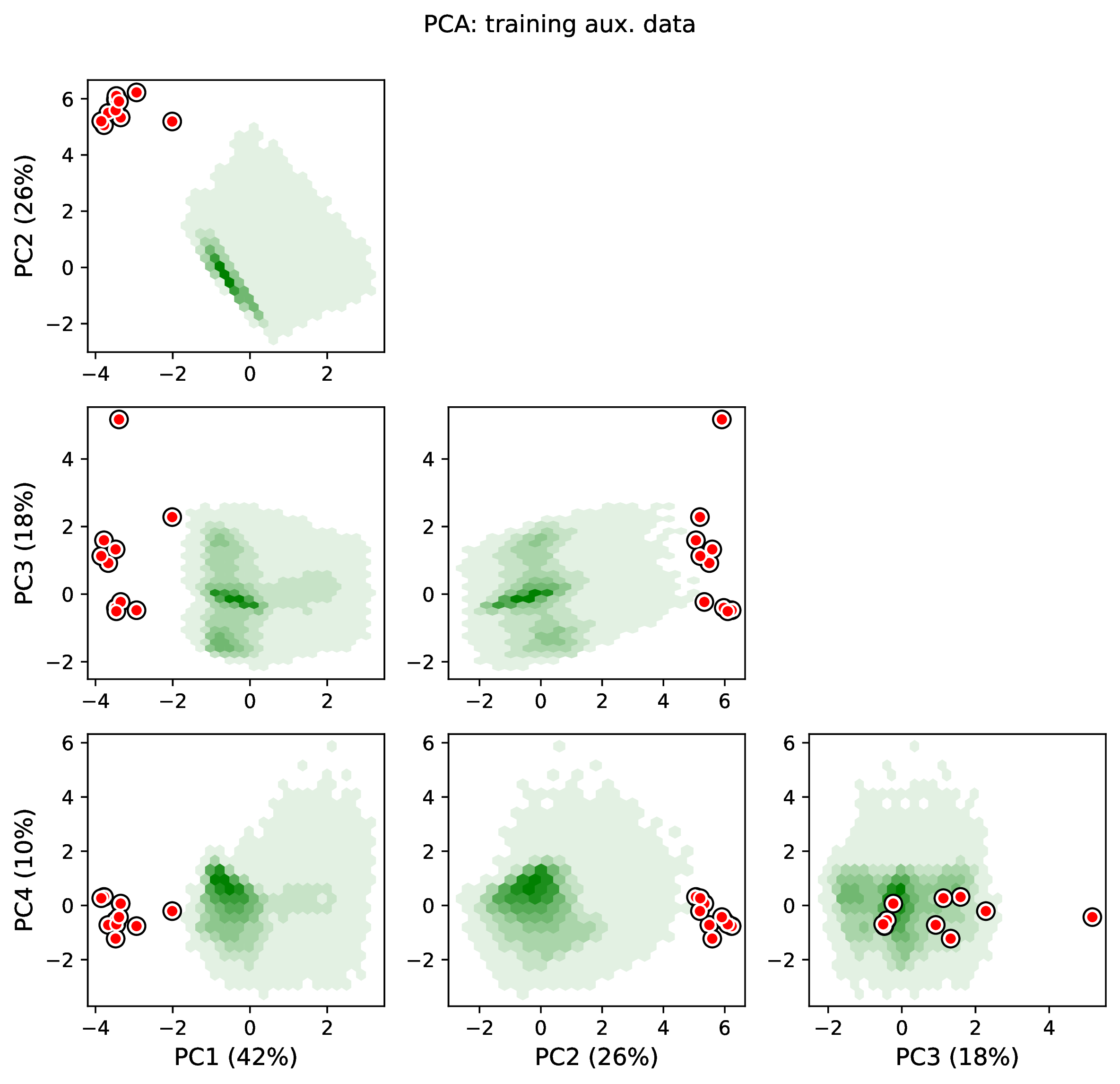
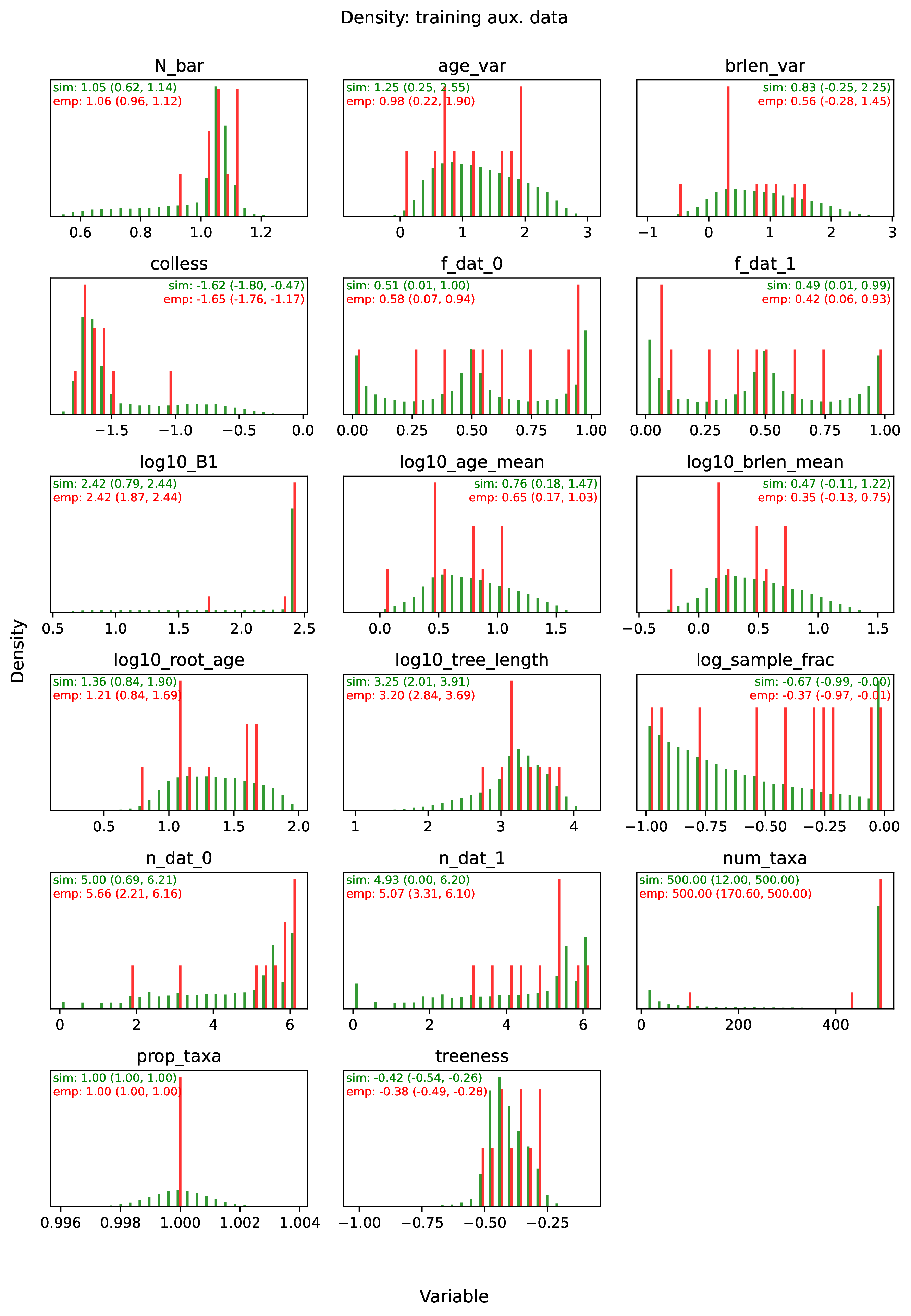
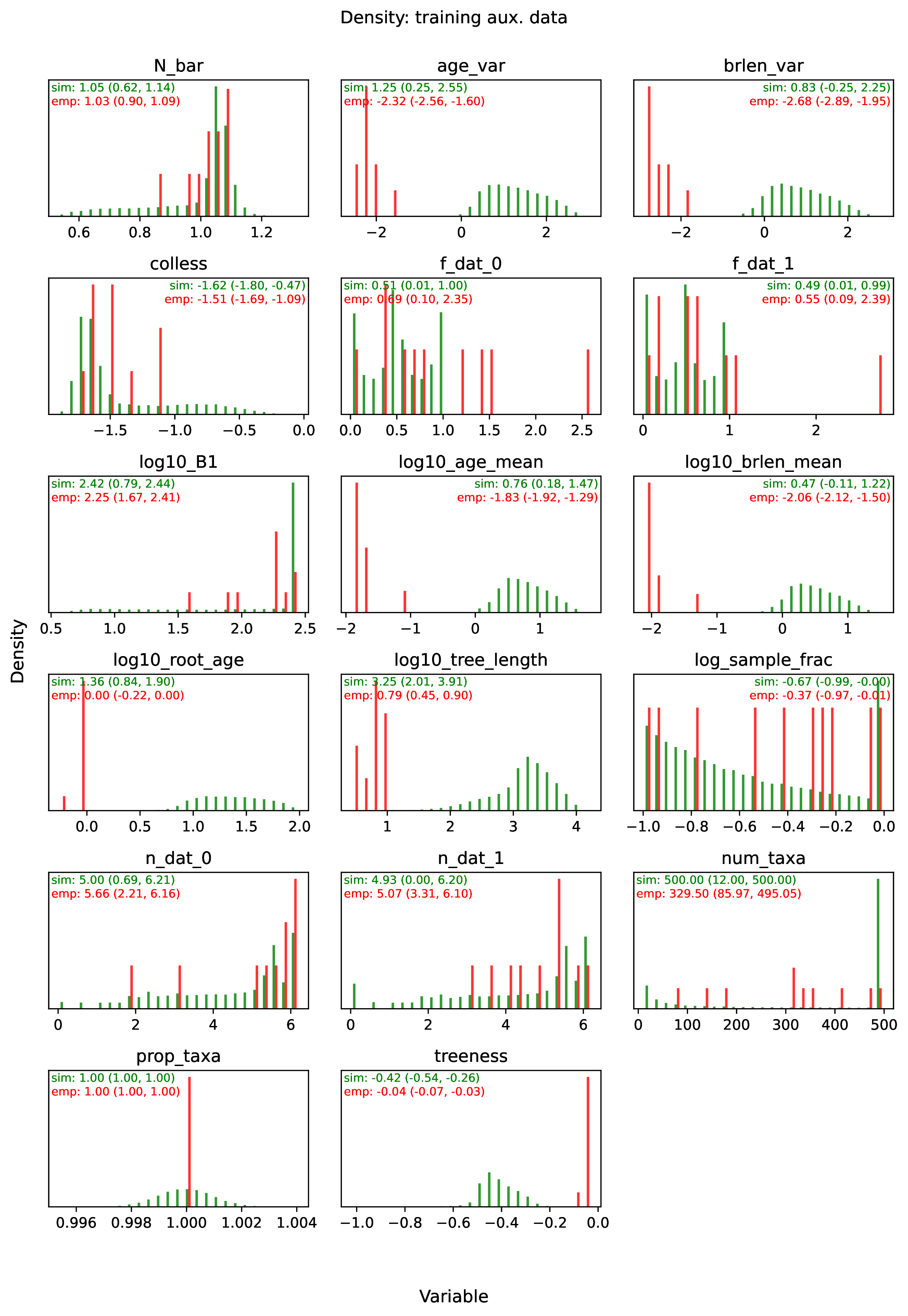
summary.pdfcontains all figures in a single plotsummary.csvrecords important results in plain text formatdensity_<dataset_name>_aux_data.pdf- densities of all values in the auxiliary dataset; red line for empirical dataset; run for training and empirical datasetsdensity_<dataset_name>_label.pdf- densities of all values in the auxiliary dataset; red line for empirical dataset; run for training and empirical datasetspca_<dataset_name>_aux_data.pdf- pairwise PCA of all values in the auxiliary dataset; red dot for empirical dataset; run for training and empirical datasetspca_<dataset_name>_label.pdf- pairwise PCA of all values in the auxiliary dataset; red dot for empirical dataset; run for training and empirical datasetstrain_history.pdf- loss performance across epochs for test/validation datasets for entire network<dataset_name>_estimate_<num_label_name>.pdf- point estimates and calibrated estimation intervals of numerical parameters for test or training datasets<dataset_name>_estimate_<cat_label_name>.pdf- confusion matrix of categorical parameters for test or training datasetempirical_estimate_num_<N>.pdf- simple plot of point estimates and calibrated prediction intervals for each empirical datasetempirical_estimate_cat_<N>.pdf- simple bar plot for each empirical datasetnetwork_architecture.pdf- visualization of Tensorflow architecture
The Tutorial and Safe Usage pages display examples of figures and explain how to interpret them.
Workspace
This section describes how phyddle organizes files and directories in its workspace.
Note
We strongly recommend that beginning users follow this general workspace filesystem design. Advanced users should find it is easy to customize locations for config files, simulation scripts, and output directories.
We recommend using the default directory structure for new projects
to simplify project management. By default, dir is set to ./ and
prefix is set to out. Output for each step is then stored in a
subdirectory named after the step and beginning with the appropriate prefix.
For example, results from Simulate are stored in the
simulate subdirectory of the dir` directory. If the sim_dir
setting is provided, Simulate results are stored into that exact
directory. For example, if dir is set to the local directory ./,
then Simulate results are saved to ./simulate. If sim_dir is
set to ../new_project/new_simulations then Simulate results are
stored there regardless of the dir setting.
Similarly, if prefix is set to out, then all
simulated datasets begin with the prefix out. If the setting sim_prefix
is set to sim, then files generated by Simulate begin with the
prefix sim.
Briefly, the workspace directory of a typical phyddle project contains
two important files
config.pythat specifies default settings for phyddle analyses in this projectsim_one.R(or a similar name) that defines a valid simulation script
and seven subdirectories for the pipeline analysis
simulatecontains raw data generated by simulationformatcontains data formatted into tensors for training networkstraincontains trained networks and diagnosticsestimatecontains new test datasets their estimationsplotcontains figures of training and validation proceduresempiricalcontains raw data for your empirical analysislogcontains runtime logs for a phyddle project
This section will assume all steps are using the example project
bundled with phyddle was generated using the command phyddle --make_cfg.
phyddle -c ./workspace/example/config.py --end_idx 25000
This corresponds to a 3-region equal-rates GeoSSE model. All directories have
the complete file set, except ./simulate contains only
20 original examples.
A standard configuration for a project named example would store pipeline
work into these directories:
./simulate # output of Simulate step
./empirical # your empirical dataset
./format # output of Format step
./train # output of Train step
./estimate # output of Estimate step
./plot # output of Plot step
./log # logs for phyddle analyses
Soon, we give an overview of the standard files and formats corresponding to each pipeline directory. First, we describe a commands that help with workspace management.
You can easily save and share your project workspace with the following command:
cd workspace/example # current directory is root of
# example project directory
phyddle --save_proj example_lite.tar.gz # save project workspace, but
# skip simulated training data
# (faster, smaller)
The resulting zip file (tarball) will contain the config file, the simulation script, and all workspace directories for pipeline steps. Note, the raw and formatted simulated training example datasets tend to be very large, and require substantial time and storage to archive, so they are not fully saved by default. To fully save all workspace project data, add the following options:
phyddle --save_proj example_full.tar.gz \ # save full project, and
--save_train_fmt T \ # include simulated training data
--save_num_sim 1000000 # (slower, larger)
If you share the project with a collaborator or save it on a server, you can load the project for use with the command:
mkdir -p ~/new_workspace/new_project # create new project directory
cd ~/new_workspace/new_project # enter new project directory
phyddle --load_proj example_lite.tar.gz # load project in directory
phyddle -s S --sim_more 10000 # (e.g.) simulate 10,000 training examples
Lastly, you can quickly remove all existing workspace directories, while preserving the config file and simulation scripts, with the following command:
cd workspace/example # enter directory to clean
phyddle --clean_proj # remove all local workspace directories
These are powerful commands, so be careful when using them. They can remove or overwrite files that you want to keep. Master these commands in a safe test directory before applying them to important workspace projects.
simulate
The Simulate step generates raw data from a simulating model that cannot yet be fed to the neural network for training. A typical simulation will produce the following files
./sim.0.tre # tree file
./sim.0.dat.csv # data file
./sim.0.labels.csv # data-generating params
Each tree file contains a simple Newick string. Each data file contains state
data either in Nexus format (.dat.nex) or simple comma-separated value format
(.dat.csv) depending on the setting for char_format.
Visit the Input datasets section to learn about the exact format for these files.
format
Applying Format to a directory of simulated datasets will output
tensors containing the entire set of training examples, stored to, e.g.
./format. If the tensor_format setting is 'csv'
(Comma-Separated Value, or CSV format), the formatted files are:
./out.empirical.phy_data.csv
./out.empirical.aux_data.csv
./out.empirical.labels.csv
./out.test.phy_data.csv
./out.test.aux_data.csv
./out.test.labels.csv
./out.train.phy_data.csv
./out.train.aux_data.csv
./out.train.labels.csv
where the phy_data.csv files contain one flattened Compact Phylogenetic Vector + States (CPV+S) entry per row, the aux_data.csv files contain one vector of auxiliary data (summary statistics and known parameters) values per row, and labels.csv contains one vector of label (estimated parameters) per row. Each row for each of the CSV files will correspond to a single, matched simulated training example. All files are stored in standard comma-separated value format, making them easily read by standard CSV-reading functions.
If the tensor_format setting is 'hdf5', the resulting files are:
./out.test.hdf5
./out.train.hdf5
./out.empirical.hdf5
where each HDF5 file contains all phylogenetic-state (CPV+S) data, auxiliary data, and label data. Individual simulated training examples share the same set of ordered examples across three internal datasets stored in the file. HDF5 format is not as easily readable as CSV format. However, phyddle uses gzip to automatically (de)compress records, which often leads to files that are over twenty times smaller than equivalent uncompressed CSV formatted tensors.
Visit Tensor formats and Auxiliary data sections to learn more about how these data are formatted.
train
Training a network creates the following files in the workspace/example/train
directory:
./out.cpi_adjustments.csv
./out.train_aux_data_norm.csv
./out.train_est.labels_cat.csv
./out.train_est.labels_num.csv
./out.train_history.csv
./out.train_label_est_nocalib.csv
./out.train_label_norm.csv
./out.train_true.labels_cat.csv
./out.train_true.labels_num.csv
./out.trained_model.pkl
Descriptions of the files are as follows, with the prefix omitted for brevity:
* trained_model.pkl: a saved file containing the trained PyTorch model
* train_label_norm.csv and train_aux_data_norm.csv: the location-scale values from the training dataset to (de)normalize the labels and auxiliary data from any dataset
* train_true.labels.csv: the true values of labels for the training and test datasets, where columns correspond to estimated labels (e.g. model parameters)
* train_est.labels.csv: the trained network estimates of labels for the training and test datasets, with calibrated prediction intervals, where columns correspond to point estimates and estimates for lower CPI and upper CPI bounds for each named label (e.g. model parameter)
* train_label_est_nocalib.csv: the trained network estimates of labels for the training and test datasets, with uncalibrated prediction intervals
* train_history.csv: the metrics across training epochs monitored during network training
* cpi_adjustments.csv: calibrated prediction interval adjustments, where columns correspond to parameters, the first row contains lower bound adjustments, and the second row contains upper bound adjustments
estimate
The Estimate step will load empirical and simulated test datasets generated by the Format step, and then make new predictions using the network trained during the Train step. Estimation will produce the following estimates, so long as the formatted input datasets can be opened in the filesystem:
./out.empirical_est.labels_num.csv # output: estimated labels for empirical data
./out.empirical_est.labels_cat.csv # output: estimated labels for empirical data
./out.test_est.labels_cat.csv # output: estimated labels for test data
./out.test_est.labels_num.csv # output: estimated labels for test data
./out.test_true.labels_cat.csv # output: true labels for test data
./out.test_true.labels_num.csv # output: true labels for test data
The out.empirical_est_labels.csv and out.test_est.labels.csv files
report the point estimates and lower and upper calibrated prediction
intervals (CPIs) for all parameters targeted by the param_est setting.
Estimates for parameters appear across columns, where columns are grouped
first by label (e.g. parameter) and then statistic (e.g. value, lower-bound,
upper-bound). For example:
$ cat out.empirical_est.labels.csv
w_0_value,w_0_lower,w_0_upper,e_0_value,e_0_lower,e_0_upper,d_0_1_value,d_0_1_lower,d_0_1_upper,b_0_1_value,b_0_1_lower,b_0_1_upper
0.2867125345651129,0.1937433853918723,0.45733220552078013,0.02445545359384659,0.002880695707341881,0.10404499205878459,0.4502031713887769,0.1966340488593367,0.5147956690178682,0.06199703190510973,0.0015074254823161301,0.27544015163806645
The test_est.labels.csv and test_true.labels.csv files contain estimated and true label values for the simulated test dataset that were left aside during training. It is crucial that estimation accuracy against the test dataset is not used to inform the training process. If you view the test results and use it to modify Train settings, you should first randomly re-sample the training and test datasets from the Format step. This helps prevent overfitting and ensures that the test dataset is truly independent of the training procedure.
plot
The Plot step generates visualizations for results previously generated by Format, Train, and (when available) Estimate.
./out.empirical_estimate_cat_{i}.pdf # categorical estimates for empirical data
./out.empirical_estimate_num_{i}.pdf # numerical estimates for empirical data
./out.test_estimate_{param} # estimation accuracy for test dataset
./out.train_estimate_{param} # estimation accuracy for training dataset
./out.train_density_aux_data.pdf # aux. data densities from Simulate/Format steps
./out.train_density_labels.pdf # label densities from Simulate/Format steps
./out.train_pca_labels_num.pdf # label PCA of Simulate/Format steps
./out.train_aux_data.pdf # aux. data PCA of Simulate/Format steps
./out.train_history_{stat}.pdf # training history for entire network
./out.network_architecture.pdf # neural network architecture
./out.summary.pdf # compiled report with all figures
./out.summary.csv # compiled text file with numerical results
The Tutorial and Safe Usage pages display examples of figures and explain how to interpret them.
empirical
The empirical directory is used to store raw data for empirical analyses.
The network from Train is only trained to make accurate predictions
for datasets with the same format as the simulate directory. That means
empirical datasets must have the same file types and formats as entries in
the simulate directory. One difference is that empirical labels.csv
files will only contain entries for “known” parameters, as specified by
param_data in the configuration; they will not contain the “unknown”
parameters to be estimated, specified by param_est.
./empirical/viburnum.0.tre # tree file
./empirical/viburnum.0.dat.csv # data file
./empirical/viburnum.0.labels.csv # data-generating params
log
The log directory contains logs for each phyddle analysis. Log files
are named according to the date and time of the analysis, and contain
runtime information that may be useful for debugging or reproducing results.
Visit Overview to learn more about the files.
Formats
This page describes different internal datatype formats and file formats used by phyddle.
Input datasets
phyddle can make phylogenetic model predictions against input datasets with
previously trained networks. Valid phyddle input datasets contain a set of
files with a shared filename prefix. For example, a dataset with the prefix
out.0 would contain a tree file out.0.tre, a character matrix file
out.0.dat.nex, and (when applicable) a ‘known parameters’ file
out.0.labels.csv. Simulated training datasets and real biological
datasets follow the same format.
Trees are encoded as raw data in simple Newick format. Trees are assumed to be rooted, bifurcating, time-calibrated trees. Trees may be ultrametric or non-ultrametric trees. Ultrametric trees should only be analyzed using treetype == ‘extant’. Non-ultrametric trees, such as those containing serially sampled viruses or fossils should be analyzed using treetype == ‘serial’. Here is an example of an extant tree with N=8 taxa.
$ cat ./simulate/out.0.tre
((((1:0.35994691486501296,2:0.35994691486501296):1.389952711060852,(3:1.5810568349100933,(4:0.5830569936279364,5:0.5830569936279364):0.9979998412821569):0.1688427910157717):5.655066077200624,6:7.404965703126489):0.3108578683347094,(7:0.7564319839861859,8:0.7564319839861859):6.959391587475013):2.2841764285388018;
Character data may be encoded in Nexus format (char_format = ‘nex’). Here is an example of a matrix with N=8 taxa and M=3 binary characters.
$ cat ./simulate/out.0.dat.nex
#NEXUS
Begin DATA;
Dimensions NTAX=8 NCHAR=3
Format MISSING=? GAP=- DATATYPE=STANDARD SYMBOLS="01";
Matrix
1 001
2 010
3 100
4 100
5 001
6 001
7 100
8 010
;
END;
Character data may also be encoded in csv format (char_format = ‘csv’). For example:
$ cat ./simulate/out.0.dat.nex
taxa,char1,char2,char3
1,0,0,1
2,0,1,0
3,1,0,0
4,1,0,0
5,0,0,1
6,0,0,1
7,1,0,0
8,0,1,0
This would represent a character matrix with 8 taxa and 3 characters with 2 states per character. phyddle will warn the user if there is a mismatch between the number of characters and states present in the character matrix and the numbers specified in the config file.
Some models will accept “known” data-generating parameters as input. For example,
if not all taxa were included in the phylogeny, a model might accept a sampling
fraction label as input. Any labels that are marked under the param_data
setting will be encoded into the auxiliary data tensor during formatting. Example:
$ cat ./simulate/out.0.labels.csv
birth_1,birth_2,death,state_rate,sample_frac
0.5728,0.9082,0.1155,0.0372,0.1114
where the setting param_data == ['sample_frac'] would ensure that only the
sample_frac entry is included in auxiliary data.
Tensor formats
Phylogenetic data (e.g. from a Newick file) and character matrix data (e.g.
from a Nexus file) are encoded into compact phylogenetic state tensors.
Internally, phyddle uses `dendropy.Tree` to represent phylogenies,
pandas.DataFrame to represent character matrices (verify), and
numpy.ndarray to store phylogenetic-state tensors.
There are two types of phylogenetic-state tensors in phyddle: the compact
bijective ladderized vector + states (CBLV+S) and the compact diversity vector +
states (CDV+S). CBLV+S is used for trees that contain serially sampled
(non-ultrametric) taxa whereas CDV+S is used for trees that contain only extant
(ultrametric) taxa. The tree_width of the encoding defines the maximum number
of taxa the phylogenetic-state tensor may contain. The tree_encode setting
determines if the tree is a 'serial' tree encoded with CBLV+S or an
'extant' tree encoded with CDV+S. Setting brlen_encode and
char_encode alter how information is stored into the
phylogenetic-state tensor.
A phyddle code data to produce the examples below is available here: link.
CBLV+S
This is an example for the CBLV+S encoding of 5 taxa with 2 characters. This is the Newick string:
(((A:2,B:1):1,(C:3,D:2):3):1,E:2);
This is the Nexus file:
#NEXUS
Begin DATA;
Dimensions NTAX=5 NCHAR=2
Format MISSING=? GAP=- DATATYPE=STANDARD SYMBOLS="01";
Matrix
A 01
B 11
C 10
D 10
E 01
;
END;
These data can be encoded in different ways, based on the char_encode
setting. When char_encode == 'integer' then the encoding will treat
each character as a row in the resulting data matrix, and assign the
appropriate integer-valued state to that character for each taxon.
Alternatively, when char_encode == 'one_hot' then the encoding will
treat every distinct state-character combination as its own row in the
resulting data matrix, then mark each species as 1 for a cell when a
species has that character-state and 0 if not. One-hot encoding is
applied individually to each homologous character (fewer distinct combinations)
not against the entire character set (more distinct combinations).
Ladderizing clades by maximum root-to-tip distance orders the taxa C, D, A,
B, then E, which correspond to the first five entries of the CBLV+S tensor.
When brlen_encode is set to 'height_only' the un-rescaled CBLV+S file
would look like this:
# NOTE: The CBLV+S tensor is shown in this orientation for readability.
# phyddle stores the tensor as the transpose of this in memory,
# meaning rows correspond to taxa, and columns correspond to branch
# length information.
# C,D,A,B,E,-,-,-,-,-
7,2,3,1,2,0,0,0,0,0 # tip-to-node distance
0,4,1,2,0,0,0,0,0,0 # node-to-root distance
1,1,0,1,0,0,0,0,0,0 # character 1
0,0,1,1,1,0,0,0,0,0 # character 2
and like this when brlen_encode is set to 'height_brlen':
# C,D,A,B,E,-,-,-,-,-
7,2,3,1,2,0,0,0,0,0 # tip-to-node distance
0,4,1,2,0,0,0,0,0,0 # node-to-root distance
3,2,2,1,2,0,0,0,0,0 # tip edge length
0,3,1,1,0,0,0,0,0,0 # node edge length
1,1,0,1,0,0,0,0,0,0 # character 1
0,0,1,1,1,0,0,0,0,0 # character 2
By default, all branch length-related entries are rescaled from 0 to 1 as proportion to tree height (formatted to ease reading):
# C, D, A, B, E, -, -, -, -, -
1.00,0.29,0.43,0.14,0.29, 0, 0, 0, 0, 0 # tip-to-node distance
0.00,0.57,0.14,0.29,0.00, 0, 0, 0, 0, 0 # node-to-root distance
0.43,0.29,0.29,0.14,0.29, 0, 0, 0, 0, 0 # tip edge length
0.00,0.43,0.14,0.14,0.00, 0, 0, 0, 0, 0 # node edge length
1, 1, 0, 1, 0, 0, 0, 0, 0, 0 # character 1
0, 0, 1, 1, 1, 0, 0, 0, 0, 0 # character 2
CDV+S
CDV+S is used to encode phylogenetic-state information for trees of only extant taxa. CDV+S has a similar structure to CBLV+S, except in two principal ways. First, CDV+S uses total subclade diversity rather than tip node with max distance-from-root-node to determine how to ladderize the tree, which in turn determines which columns are associated with which tip nodes. Second, because CDV+S is used for extant-only trees, it does not need to report the redundant information about tip-to-node distances, as the tip-to-root distances are equal among all tips (by definition). This means that CDV+S does not contain a row with tip-to-node distances (the first row of CBLV+S).
For example, the following Newick string for an ultrametric tree
(((A:5,B:5):1,(C:3,D:3):3):1,E:7);
and associating the same character data as above with taxa A through E yields the following CDV+S tensor:
# NOTE: The CDV+S tensor is shown in this orientation for readability.
# phyddle stores the tensor as the transpose of this in memory,
# meaning rows correspond to taxa, and columns correspond to branch
# length information.
# B,A,C,D,E,-,-,-,-,-
2,1,4,0,0,0,0,0,0,0 # node-to-root distance
5,5,3,3,7,0,0,0,0,0 # tip edge length
1,1,3,0,0,0,0,0,0,0 # node edge length
1,0,1,1,0,0,0,0,0,0 # character 1
1,1,0,0,1,0,0,0,0,0 # character 2
By default, all branch length-related entries are rescaled from 0 to 1 as proportion to tree height (formatted to ease reading):
# B, A, C, D, E,-,-,-,-,-
0.29,0.14,0.57,0.00,0.00,0,0,0,0,0 # node-to-root distance
0.71,0.71,0.43,0.43,1.00,0,0,0,0,0 # tip edge length
0.14,0.14,0.43,0.00,0.00,0,0,0,0,0 # node edge length
1, 0, 1, 1, 0,0,0,0,0,0 # character 1
1, 1, 0, 0, 1,0,0,0,0,0 # character 2
Auxiliary data
The auxiliary data tensor contains a panel of summary statistics extracted from the inputted phylogeny and character data matrix for a given dataset: Currently, phyddle generates the following summary statistics:
log10_tree_length # sum of branch lengths
log10_root_age # longest root-to-tip distance
log10_brlen_mean # mean of branch lengths
log10_age_mean # mean of internal node ages
age_var # variance of internal node ages
brlen_var # variance of branch lengths
log10_B1 # B1 tree measure (Dendropy)
colless # Colless tree measure (Dendropy)
treeness # treeness measure (Dendropy)
N_bar # N_bar tree measure (Dendropy)
f_dat_0 # frequency of taxa with character in state 0
f_dat_1 # frequency of taxa with character in state 1
n_dat_0 # count of taxa with character in state 0
n_dat_1 # count of taxa with character in state 0
num_taxa # number of terminal taxa in tree/data
prop_taxa # proportion of taxa retained after downsampling to tree_width
...
The auxiliary data tensor can also contain specified parameter values that shape the
data-generating process and can be treated as “known” rather than needing to
be estimated. For example, the epidemiologists may assume they know the rate of
infection recovery (gamma) based on public health or clinical data. Parameters
may be treated as data by providing the labels for those parameters in the
param_data entry of the config file. For example, setting 'param_data' :
[ 'sample_frac' ] could be used to inform phyddle that the recovery
rate and susceptible population sizes for location 0 are known for a
phylogenetic SIR analysis.
Safe Usage
We designed phyddle to be user-friendly. However, we caution users that deep learning approaches can easily produce nonsensical and/or misleading results when irresponsibly. Below, we describe several issues that newcomers to deep learning might encounter, along with how to diagnose and solve them.
Poor accuracy or coverage
Neural networks are trained to make accurate predictions, such as point estimates and prediction intervals. Poor accuracy or coverage is a common problem and easily detected, but it can arise for many reasons.
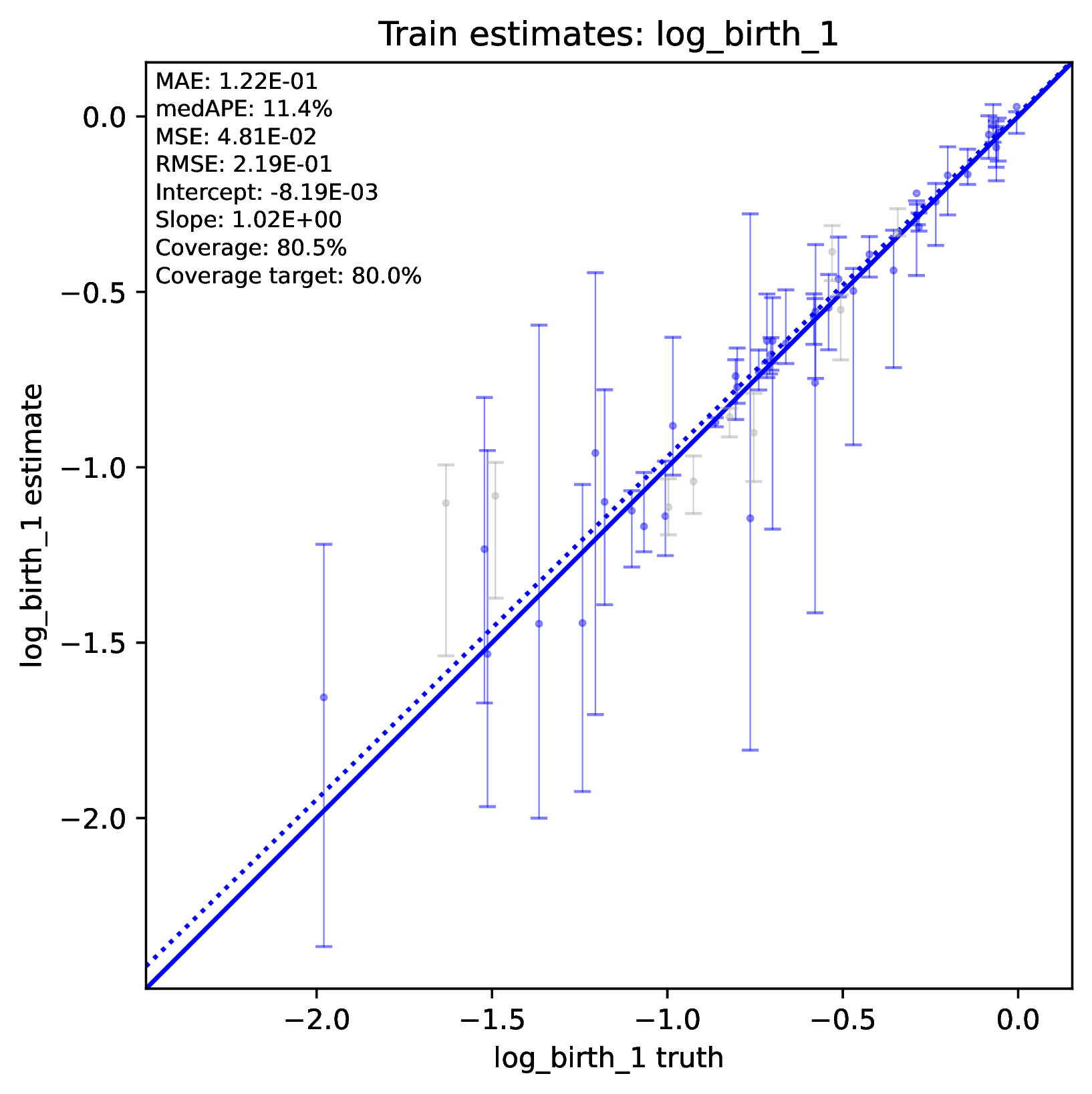
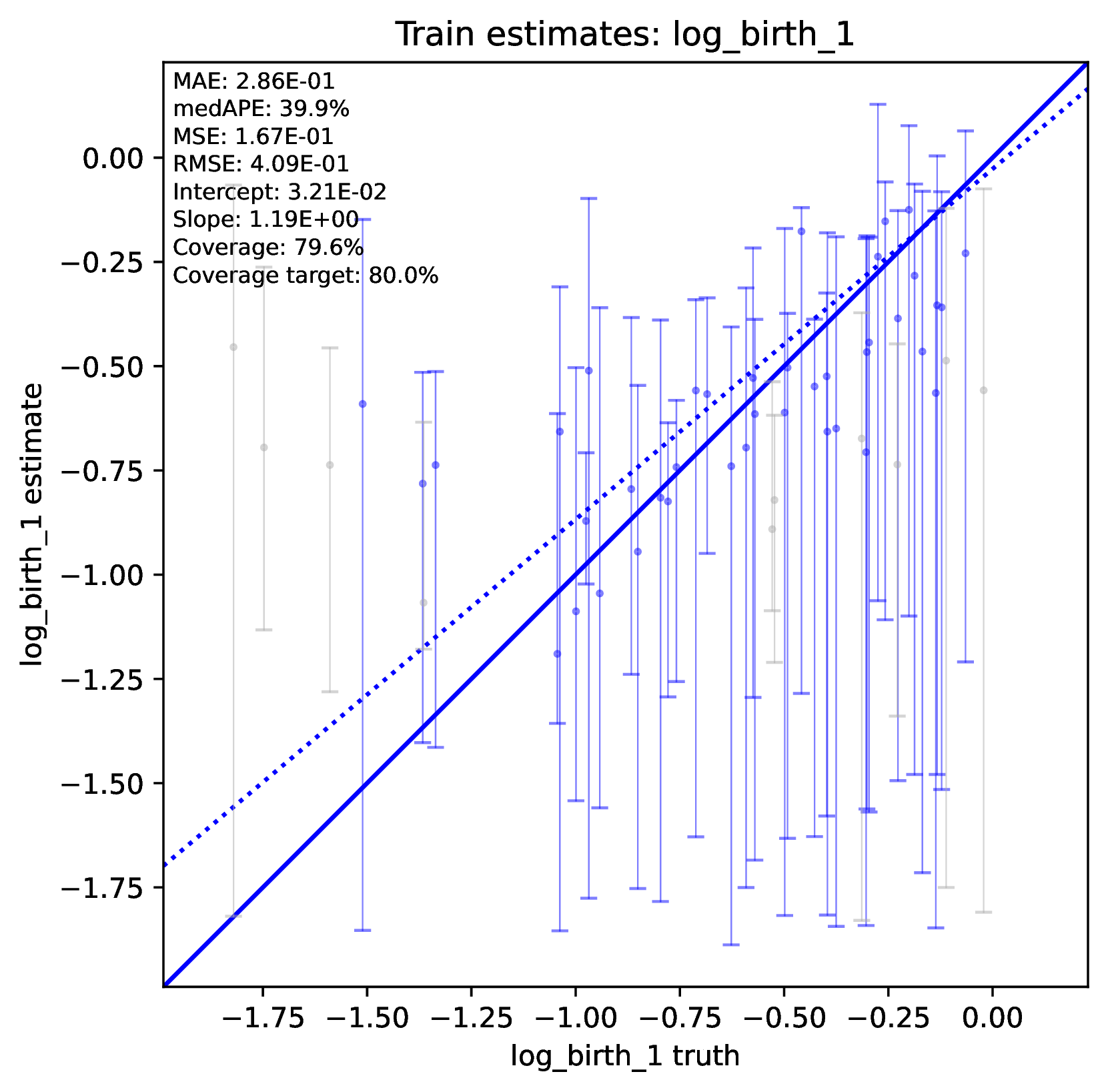
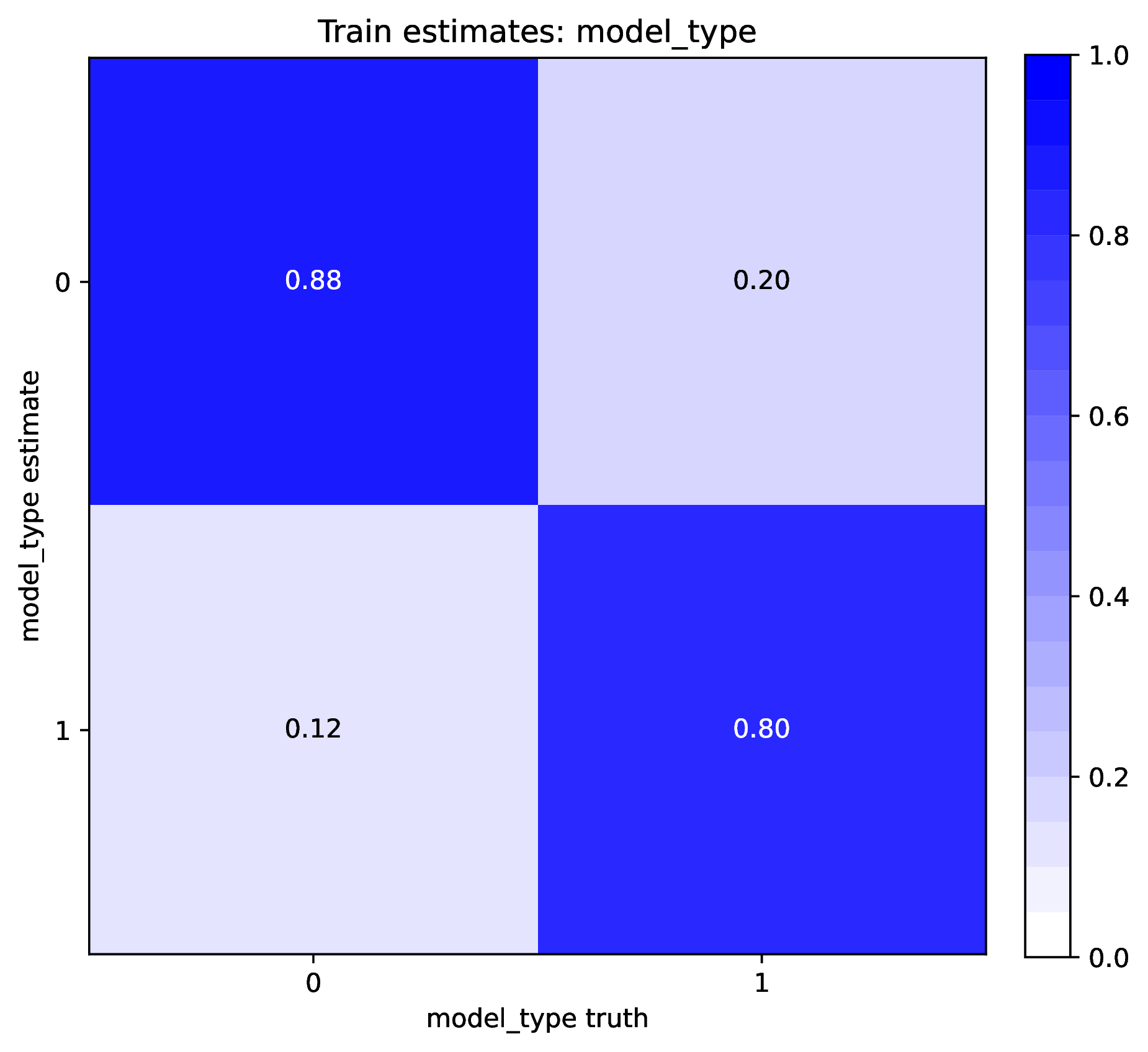
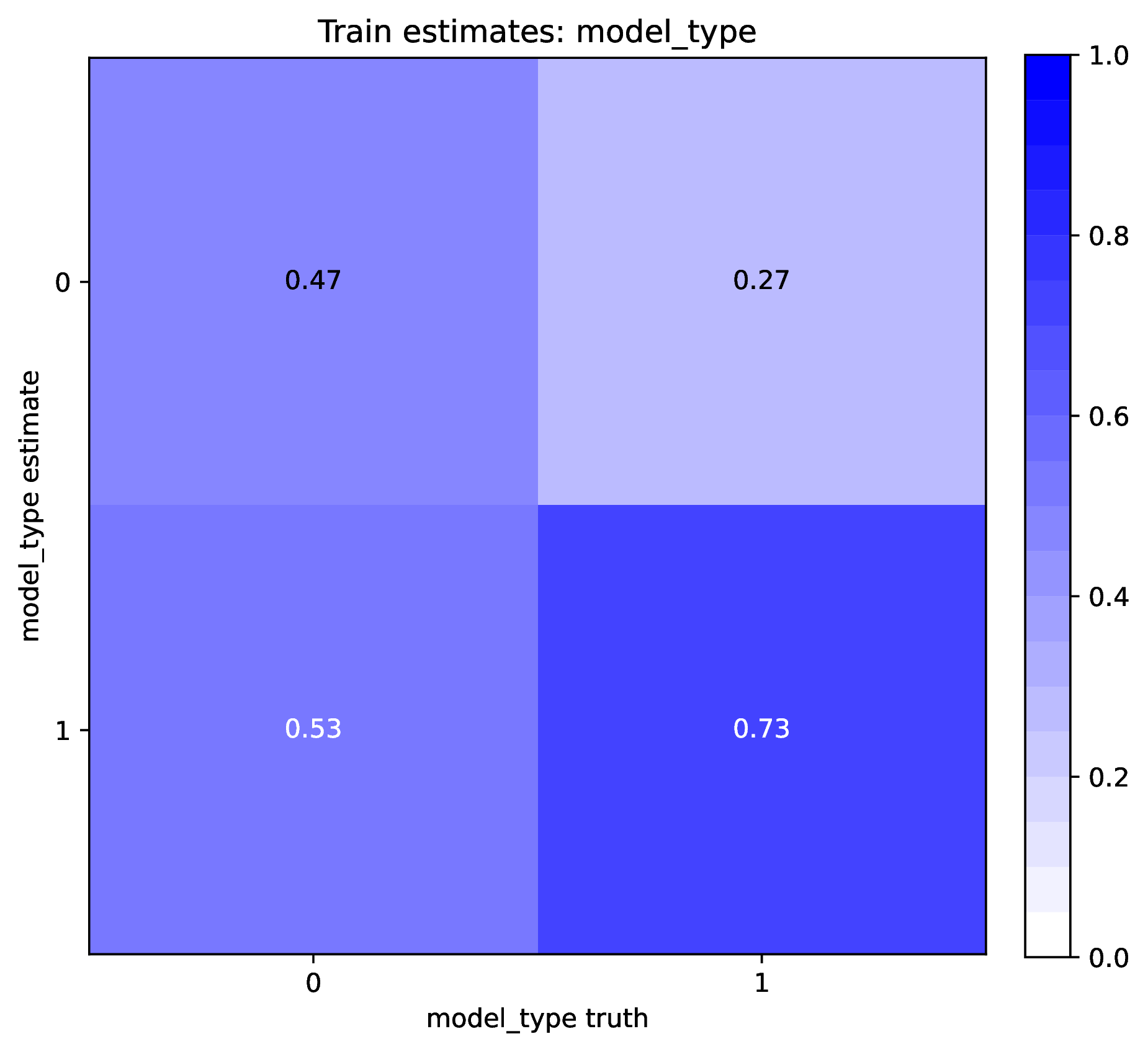
To diagnose poor accuracy or coverage, run the Plot step. Review the scatter plots of true versus estimated labels for the Train and Test datasets. Accurate predictions for numerical labels should fall along the 1:1 line, with a slope close to 1.0. Coverage levels should be close to the target level. For categorical variables, the confusion matrix should show high values along the diagonal and small values elsewhere. phyddle will print warnings if accuracy or coverage is poor in the Training or Test datasets.
To correct the issue, undertraining, overtraining, small training datasets, poor network architecture, or other issues may be at play. The following sections provide ideas to fix the issue.
Undertraining and overtraining
The loss score for the training dataset will typically decrease as the number of training epochs increases. This is because typically neural networks have thousands of weight and bias parameters to adjust, and small adjustments can easily squeeze at marginal improvements to the loss score.
An undertrained network will not make accurate predictions for training, test, or validation datasets. This is because the optimizer was not able to learn the optimal network parameters before training concluded. On the other hand, an overtrained network will only make accurate predictions for the training dataset, while making inaccurate predictions against non-training datasets, such as the test and validation datasets.
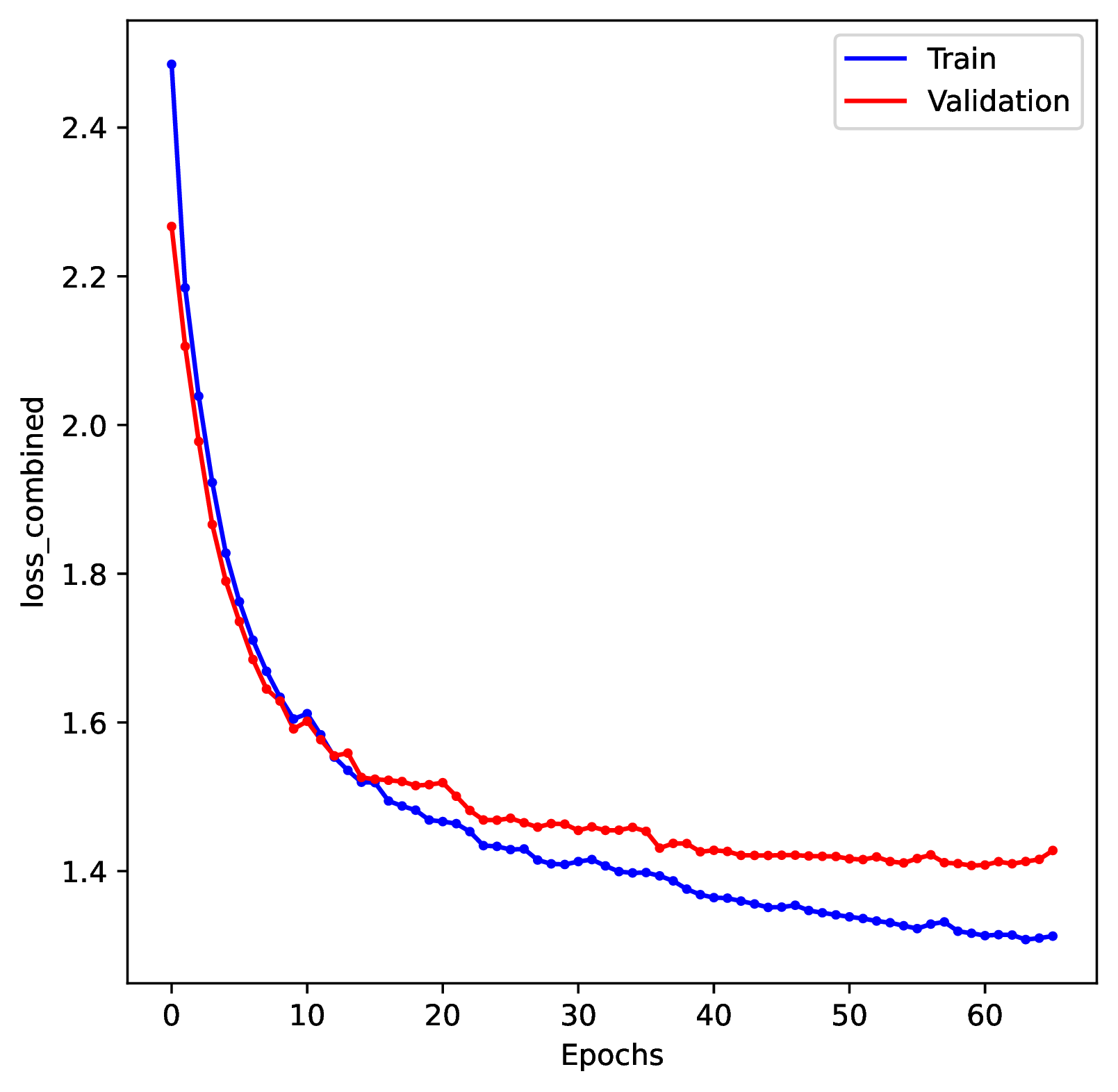
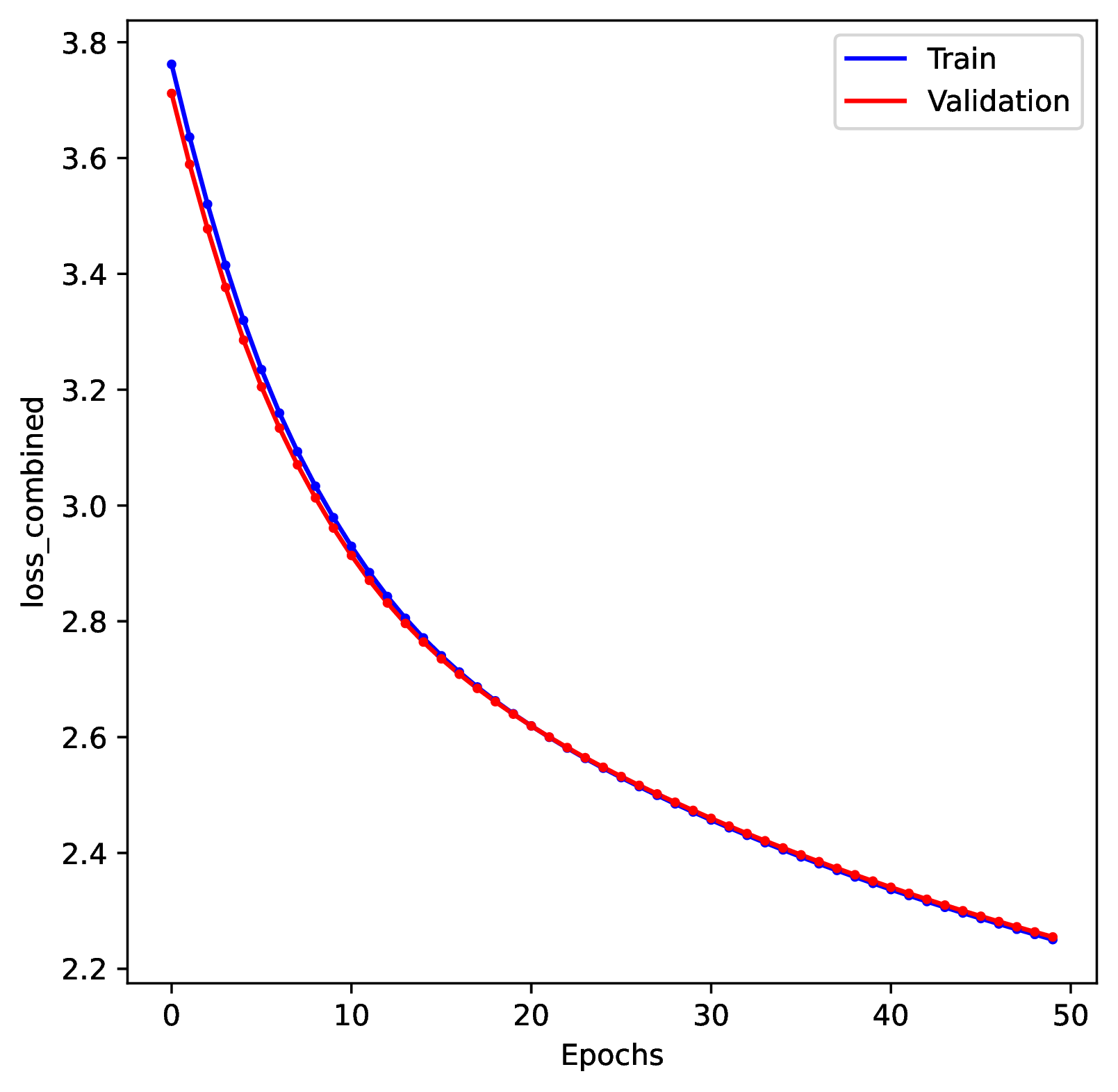
To diagnose undertraining problems, review the training history plot. If the loss score for both the training and validation datasets is still decreasing by the end of the training period, then the network is undertrained.
To correct for undertraining, double the number of training epochs or use larger batch sizes of training examples.
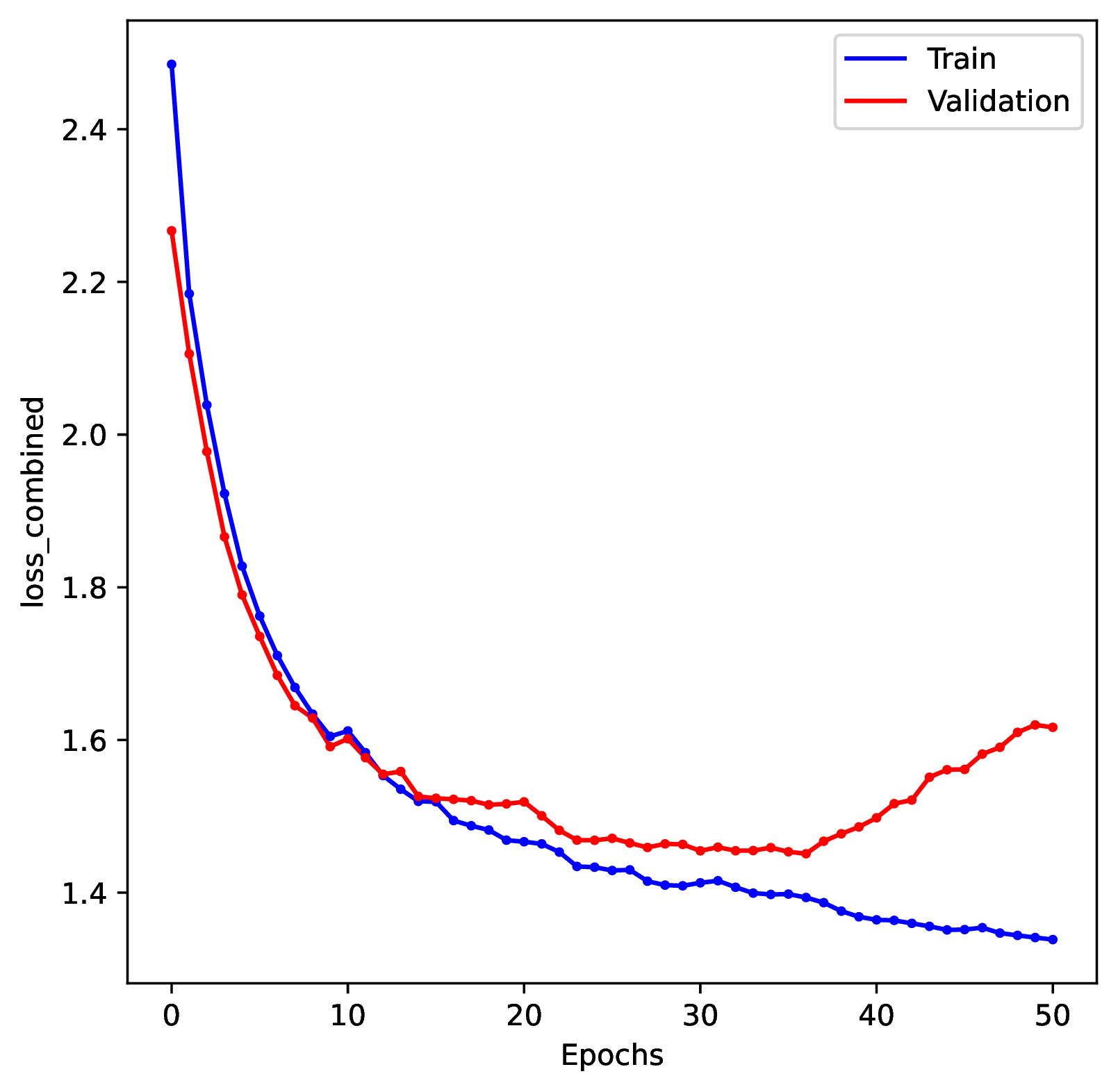
To diagnose overtraining problems, run the Plot step. Review the training history plot. If you see that the loss score for the training dataset continuously decreases while the loss score for the validation dataset increases (making a ‘U’ shape) then the network is overtrained.
To correct for overtraining, you can either specify a shorter number of training epochs, decrease the training batch sizes, or use stronger early stopping rules. To prevent overtraining, the default behavior in phyddle is to stop training when the validation loss score increases for 3 consecutive epochs.
Small training datasets
Supervised learning requires large training datasets to train networks to perform accurately. Most of the workspace examples are designed to work well with ~50,0000 training examples. Small training datasets do not contain enough examples to densly represent general relationships between input and output patterns. In this case, the trained network will have poor prediction accuracy against the training, test, and validation datasets – even if the network was trained to its optimal level.
To diagnose this issue, run the Plot step. Verify that the training history plot does not show evidence for overtraining or undertraining. Next, verify the network has poor prediction accuracy for the training and test datasets.
To fix the issue, simulate twice as many datasets, retrain the network, and then assess whether accuracy improved. If it does, continue simulating data until prediction accuracy for the training and test datasets stabilizes.
Suboptimal network size
Neural networks are composed of layers of nodes that are interconnected through series of activation functions that are optimized during training. In general, increasing network depth (more layers) and width (more nodes) allows a network to easily express a broader variety of relationships between inputs and outputs (higher capacity), which can increase accuracy when properly trained. However, because larger networks have more parameters to optimize, they are also prone to overtraining.
To diagnose issues with network size, run the Plot step. If the network is too small, the network will underperform across all prediction tasks. The training history plot will show rapid convergence towards the optimal loss score, but the final loss score will be higher than expected. In addition, prediction accuracy for the training and test datasets will be in agreement. If the network is too large, then the training history will show evidence of overtraining, and the prediction accuracy for the training dataset will be high, with degraded accuracy for the test dataset.
To fix the issue, compare the prediction accuracy of networks trained with different depths and widths. To do this, modify the numbers of layers and nodes through the configuration file, train the new network, and compare differences in accuracy and training time with other network designs.
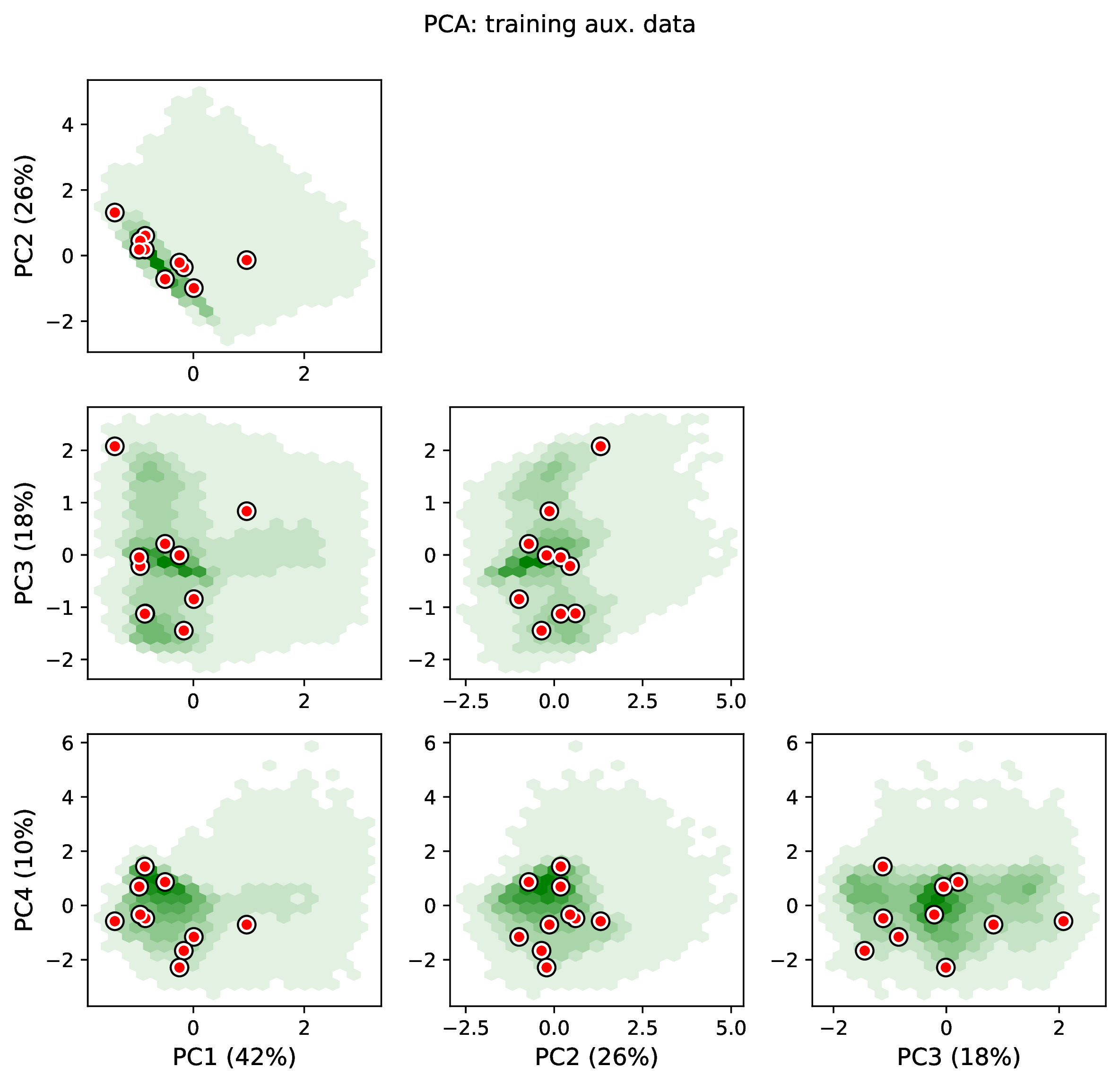
Out-of-distribution examples
Supervised learning uses large datasets, composed of pairs of input (features) and output (labels), to train neural networks to accurately predict labels from new datasets. Properly trained network should perform well at making predictions for inputs similar to those in the training dataset. For example, the training, test, validation, and calibration examples are all samples from the same distribution of data (i.e. through the model-based simulator). New empirical datasets might look like these data (in-distribution) or they might look radically different (out-of-distribution). Out-of-distribution examples are do not resemble any training examples, so the network cannot be trusted to make accurate predictions from them.
To diagnose this issue, run the Estimate and Plot steps. The Estimate step will report any empirical datasets and variables that are outliers with respect to the training dataset. The Plot step will generate PCA plots and histograms of variables for the training and empirical datasets; inspect the plots to identify empirical examples that are outliers.
To solve the problem, verify the empirical datasets are formatted in the same way as the training dataset. For example, make sure phylogenetic branch lengths and traits are measured in the same units, and that discrete trait labels use the same encoding. If they are formatted correctly, then you will need to simulate a new and more-variable training dataset that will include the range of expected input and output variables from the empirical data. For example, if your empirical trees are roughly 20 to 50 million years in height, consider simulating training trees that are between 5 and 100 million years in height.
Simulator errors
phyddle uses simulated examples to train neural networks. This means that the quality of the training data is only as good as the simulator that generates it. Untested simulators often contain coding or mathematical errors. We stress that if the simulator generates erroneous datasets, then the network will be trained to confidently make erroneous predictions!
Diagnosing the issue is not easy. A network trained on a bad simulator will still make accurate predictions for the training, test, and validation datasets. However, predictions for empirical datasets will remain bad despite attempts to adjust the size or variability of training examples, network architecture, or training settings.
Use other simulators, estimation methods, and theory to validate the correctness of your simulator. One good option is to find a special case of your model that is identical to a second more-tractable model: e.g. reduce your time-heterogeneous SSE model to a pure birth process. In these cases, you can compare the output of your simulator to the output from a second peer-reviewed simulator. Simulate data with both simulators under the same model conditions, and verify they produce similar distributions of output: e.g. both simulators yield trees with similar tree height distributions. Another option is to use or derive theory to verify that your simulator produces the expected output: e.g. show your simulator generates the correct expected number of species for a given set of parameters and duration of evolutionary time. In cases where your model has a tractable likelihood, the estimates of your trained network should agree with those from a likelihood-based estimator. Likelihood-based Bayesian coverage experiments for extremely small datasets (e.g. 3 species, 2 characters) are sufficient to show the simulator is correct.
Unintended signal
Neural networks are designed to learn patterns from data, which means they can also learn unintended patterns. In particular, phyddle networks are only expected to make accurate predictions for new empirical datasets that follow the same format as the simulated datasets. Unless handled carefully by the analyst, simulated and empirical datasets might differ in terms of scale (e.g. thousands vs. billions of years for tree height), transformation (e.g. log vs. linear trait measurements), character encoding (e.g. the simulator sorts nucleotide sites by variability, but the empirical preserves its ordering), state encoding (e.g. the simulator uses state 0, but the empirical data uses state 1 to represent a widespread species in regions A and B), or other ways. While phyddle will internally rescale traits and tree heights internally to mitigate some issues, it cannot automatically correct for biases that might arise due to encodings or transformations.
To diagnose the issue, run the Estimate and Plot steps with the empirical data included. If the data representation is grossly mismatched, the issue may appear as an out-of-distribution error, and you may see that the empirical data are outliers in the PCA plots and/or histograms. Identifying the issue for subtle differences in data representation may require a deep understanding of the model and the data.
To correct the issue, you need to ensure that the empirical data are represented in the same way as the simulated data. For example, if your simulator log-transforms traits before saving the simulation to file, you must do the same for your empirical dataset. A good strategy for this is to write code for necessary data adjustments (e.g. log-transformations, shuffling character order) and that same code for both the simulated and empirical data.
Model design
There are many subtle factors related to model design that can impact the performance of estimation tasks. For example, models with non-identifiable parameters can be difficult or impossible to estimate – e.g. estimating compound parameters for evolutionary distance rather than separate parameters for rate and time. In other cases, the parameterization of the model can impact learning efficiency – e.g. using birth and death rates versus diversification and turnover parameters is known to influence how easily maximum likelihood methods estimate birth-death model parameters.
There are far too many issues related to model design to describe here. In general, it is safe to assume that deep learning methods will face similar difficulties as likelihood-based methods for standard model estimation tasks. If theory shows that a model is non-identifiable, neither method would precisely estimate all of that model’s parameters.
Designing models is complicated and benefits from mathematical expertise and experience. We recommend that biologists who are creating novel models for the first time consult with theoreticians identify potential flaws in the model design.
Nonsensical results
When applying a statistical model or inference method to analyze an empirical dataset, it’s critical to verify that the results are sensible. Models and methods cannot put the results it finds into a broader scientific context. For example, inferring a net diversification rate of 1,000 species per species per Myr for a bird clade might be an earth-shattering result that fundamentally changes our understanding of vertebrate evolution. Or perhaps the model is poorly designed. Or perhaps the simulator contained a bug. Or perhaps the network was overtrained. Or perhaps the dataset contained an error.
We urge biologists to use their knowledge of the empirical system and the model properties to interpret results. Do not hesitate to question the results of any statistical analysis, with phyddle or otherwise, especially if they cannot be explained clearly and convincingly. If you suspect your results cannot be trusted, explore whether results are robust to adjustments in the dataset or analysis settings, and to what extent the results can be replicated with other methods.
Assorted tricks
Here are a few tricks for using phyddle using a Unix-based terminal. These commands assume a standard phyddle workspace directory structure.
Make a new config file
# Create and modify new config file
phyddle --make_cfg
# Rename new config
mv config_default.py my_new_config.py
# Design new config
edit my_new_config.py
# Run using new config
phyddle -c my_new_config.py
Run a pipeline with modified command-line settings
# Run full pipeline while changing calibration and validation proportions
phyddle -c config.py --cal_prop 0.10 --val_prop 0.10
Re-run part of the pipeline with modified command-line settings
# Re-run pipeline Train, Estimate, and Plot steps with new training settings
phyddle -c config.py -s TEP --num_epoch 10 --trn_batch_size 64
Redirect input/output across pipeline steps
# Run full pipeline
phyddle -c config.py
# Re-run Train, Estimate, Plot steps with new settings, saved to other_project
phyddle -c config.py \
-s TEP \
--trn_dir ../other_project/train \
--est_dir ../other_project/estimate \
--plt_dir ../other_project/plot \
--num_epochs 40 \
--trn_batch_size 512
# Alternatively, point `dir` to other project point Format to this project
phyddle -c config.py \
-s TEP \
--dir ../other_project \
--fmt_dir ./format \
--num_epochs 40 \
--trn_batch_size 512
Process alternative empirical data using previously trained network
# Run full pipeline
phyddle -c config.py
# Format, Estimate, and Plot empirical data against trained network
phyddle -c config.py \
-s FEP \
--no_sim \
--emp_dir ../other_project/empirical \
--est_dir ./new_estimate \
--plt_dir ./new_plot
Simulate new training examples
# Simulate training examples 0 to 999, storing results
# workspace/simulate/my_project
phyddle -s S -c config.py --start_idx 0 --end_idx 1000
# Simulate 4000 more training examples, 0 to 4999
phyddle -s S -c config.py --sim_more 4000
# Perform remaining Format, Train, Estimate, Plot steps
phyddle -s FTEP -c config.py
# ...or, to Simulate more and re-run all steps
phyddle -c config.py --sim_more 4000
Run an analysis for a tree without character data
To analyze a model that only generates trees but not character data (e.g. a birth-death process), design your simulator to write to file a character matrix with the same taxon set, but with zeroes for all character state information. When training, only the trees will contain variation that can be used to estimate model parameters. Future versions of phyddle will allow for the direct analysis of trees without character data, without needing this workaround.
Run an analysis for data without a tree
To analyze a model that only generates data but no tree (e.g. a normal
distribution), design your simulator to write a two-taxon tree
((A:1,B:1);) to the tree file, a two-taxon character matrix with zeroes
for states to the character matrix file (see above), and all data of actual
interest into the labels file. Then, identify all variables of interst in
param_data. When training, only the known parameters will contain
variation that can be used to estimate model parameters. Future versions of
phyddle will allow for the direct analysis of data without trees and
character data, without needing this workaround.
Quick access to workspace directories from console via GUI
On Mac OS X, you can press the Option key and click a directory path to open a Finder window to that directory.
Example run
The output of phyddle pipeline analysis will resemble this:
┏━━━━━━━━━━━━━━━━━━━━━━┓
┃ phyddle v0.1.1 ┃
┣━━━━━━━━━━━━━━━━━━━━━━┫
┃ ┃
┗━┳━▪ Simulating... ▪━━┛
┃
┗━━━▪ output: ./simulate
▪ Start time of 20:37:07
▪ Simulating raw data
Simulating: 100%|█████████████████████████| 100/100 [00:20<00:00, 4.94it/s]
▪ Total counts of simulated files:
▪ 31030 phylogeny files
▪ 31030 data files
▪ 31030 labels files
▪ End time of 20:37:31 (+00:00:24)
... done!
┃ ┃
┗━┳━▪ Formatting... ▪━━┛
┃
┣━━━▪ input: ./empirical
┃ ./simulate
┗━━━▪ output: ./format
▪ Start time of 20:37:32
▪ Collecting simulated data
▪ Encoding simulated data as tensors
Encoding: 100%|██████████████████████| 31030/31030 [04:36<00:00, 112.08it/s]
Encoding found 31030 of 31030 valid examples.
▪ Splitting into train and test datasets
▪ Combining and writing simulated data as tensors
Making train hdf5 dataset: 29479 examples for tree width = 500
Making test hdf5 dataset: 1551 examples for tree width = 500
▪ Collecting empirical data
▪ Encoding empirical data as tensors
Encoding: 100%|█████████████████████████████| 10/10 [00:50<00:00, 5.08s/it]
Encoding found 10 of 10 valid examples.
▪ Combining and writing empirical data as tensors
Making empirical hdf5 dataset: 10 examples for tree width = 500
▪ End time of 20:43:21 (+00:05:49)
... done!
┗━┳━▪ Training... ▪━━┛
┃
┣━━━▪ input: ./format
┗━━━▪ output: ./train
▪ Start time of 20:45:09
▪ Loading input:
▪ 22111 training examples
▪ 5895 calibration examples
▪ 1473 validation examples
▪ Training targets:
▪ log10_birth_1 [type: num]
▪ log10_birth_2 [type: num]
▪ log10_death [type: num]
▪ log10_state_rate [type: num]
▪ Building network
▪ Training network
Training epoch 1 of 10: 100%|███████████████| 44/44 [00:50<00:00, 1.14s/it]
Train -- loss: 1.0648
Validation -- loss: 0.9459
Training epoch 2 of 10: 100%|███████████████| 44/44 [00:49<00:00, 1.13s/it]
Train -- loss: 0.8429 abs: -0.2219 rel: -20.84%
Validation -- loss: 0.8125 abs: -0.1333 rel: -14.10%
Training epoch 3 of 10: 100%|███████████████| 44/44 [00:49<00:00, 1.12s/it]
Train -- loss: 0.7646 abs: -0.0782 rel: -9.28%
Validation -- loss: 0.7716 abs: -0.0410 rel: -5.04%
Training epoch 4 of 10: 100%|███████████████| 44/44 [00:49<00:00, 1.12s/it]
Train -- loss: 0.7218 abs: -0.0429 rel: -5.61%
Validation -- loss: 0.7275 abs: -0.0441 rel: -5.71%
Training epoch 5 of 10: 100%|███████████████| 44/44 [00:48<00:00, 1.11s/it]
Train -- loss: 0.6917 abs: -0.0300 rel: -4.16%
Validation -- loss: 0.6930 abs: -0.0345 rel: -4.74%
Training epoch 6 of 10: 100%|███████████████| 44/44 [00:48<00:00, 1.11s/it]
Train -- loss: 0.6657 abs: -0.0261 rel: -3.77%
Validation -- loss: 0.6874 abs: -0.0056 rel: -0.81%
Training epoch 7 of 10: 100%|███████████████| 44/44 [00:49<00:00, 1.13s/it]
Train -- loss: 0.6417 abs: -0.0240 rel: -3.61%
Validation -- loss: 0.6621 abs: -0.0253 rel: -3.68%
Training epoch 8 of 10: 100%|███████████████| 44/44 [00:50<00:00, 1.14s/it]
Train -- loss: 0.6264 abs: -0.0153 rel: -2.38%
Validation -- loss: 0.6549 abs: -0.0072 rel: -1.09%
Training epoch 9 of 10: 100%|███████████████| 44/44 [00:49<00:00, 1.13s/it]
Train -- loss: 0.6144 abs: -0.0119 rel: -1.91%
Validation -- loss: 0.6376 abs: -0.0173 rel: -2.64%
Training epoch 10 of 10: 100%|██████████████| 44/44 [00:49<00:00, 1.14s/it]
Train -- loss: 0.6078 abs: -0.0067 rel: -1.08%
Validation -- loss: 0.6307 abs: -0.0069 rel: -1.08%
▪ Processing results
▪ Saving results
▪ End time of 20:53:47 (+00:08:38)
▪ ... done!
┃ ┃
┗━┳━▪ Estimating... ▪━━┛
┃
┣━━━▪ input: ./format
┃ ./train
┗━━━▪ output: ./estimate
▪ Start time of 20:53:47
▪ Estimation targets:
▪ log10_birth_1 [type: num]
▪ log10_birth_2 [type: num]
▪ log10_death [type: num]
▪ log10_state_rate [type: num]
▪ Loading simulated test input
▪ Making simulated test estimates
▪ Loading empirical input
▪ Making empirical estimates
▪ End time of 20:53:49 (+00:00:02)
... done!
┃ ┃
┗━┳━▪ Plotting... ▪━━┛
┃
┣━━━▪ input: ./format
┃ ./train
┃ ./estimate
┗━━━▪ output: ./plot
▪ Start time of 20:55:18
▪ Loading input
▪ Generating individual plots
▪ Combining plots
▪ Making csv report
▪ End time of 20:55:35 (+00:00:17)
... done!